The DNA replication group at the LMS was formed in 2006. The team, led by Christian Speck, researches fundamental principles in DNA replication, and how this process contributes to chromatin organisation, genome architecture and transcriptional regulation. We use this knowledge to understand the molecular basis of disease and to develop novel cancer therapies.
“How does DNA replication start and how is it controlled?”
DNA replication requires complete control and a high level of fidelity – incorrectly replicating DNA can have disastrous results, ranging from cell death to disease.
The helicase is an enzyme essential in DNA replication. It “unwinds” the DNA strands so replication can begin. A set of proteins, origin recognition complex 1-6, helps to guide the helicase to its correct starting position. Not much is known about how DNA replication sites are chosen or how access by the helicase is controlled.
Our research focuses on these replication factors. We want to find out how they function and how they help to organise DNA into tightly packed and loose sections, which in turn regulate gene expression.
We use a range of interdisciplinary approaches, including biochemistry, high-resolution genomics, genome-wide AlphaFold predictions, and proteomics.
We also use cryogenic electron microscopy, where samples are cooled to very low temperatures to preserve their structures before being studied under the microscope.
With a better understanding of these replication factors, we hope we can find out more about disease-associated processes and develop novel DNA replication inhibitors that could help treat cancers.
“We study how DNA replication is controlled, how this process becomes misregulated in disease and use our insights to develop novel DNA replication inhibitors.”
The objective of Speck Lab is to discover new mechanisms in initiation of DNA replication and to understand the function of replication factors in heterochromatin formation and epigenetic memory. This knowledge is used to understand disease-associated processes and in order to develop novel DNA replication inhibitors. The team employs truly interdisciplinary approaches including biochemistry, cryo-EM, high resolution genomics, chemical biology, genetics and molecular dynamics simulations to obtain holistic understanding of biological processes.
To find out more visit Specklab.com and the group leader’s Imperial College website.
Our team studies how DNA replication is controlled, because this process is essential for maintaining genome stability and supporting healthy ageing. When replication goes wrong, it can contribute to rare conditions such as Meier-Gorlin Syndrome and one of the most common health problems, cancer.
By combining cutting-edge computational tools, biochemistry, and genomics, we are uncovering novel regulators of DNA replication and revealing how mutations in these proteins can lead to disease. We also work with collaborators to develop new small molecules and peptides that block DNA replication, with the goal of creating new cancer therapies. In addition, the team studies how DNA replication factors influence telomeres, discovering new regulatory circuits that are misregulated in cancer.


Reuter LM, Khadayate SP, Mossler A, Liebl K, Faull SV, Karimi MM, Speck C. (2024). MCM2-7 loading-dependent ORC release ensures genome-wide origin licensing. Nat Commun. 2024 Aug 24;15(1):7306. doi: 10.1038/s41467-024-51538-9
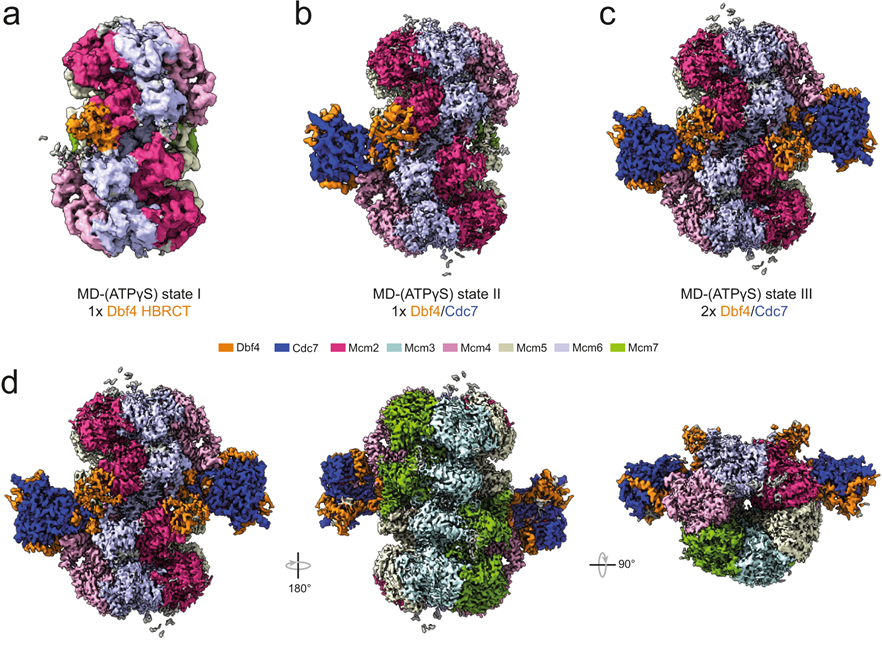
Saleh A., Noguchi Y., Aramayo R., Ivanova ME., Stevens KM., Montoya A., Sunidhi S., Carranza NL., Skwark MJ., Speck C. (2022). The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer. Nat Commun 13, 2915. https://doi.org/10.1038/s41467-022-30576-1
Feng X., Noguchi Y., Barbon M., Stillman B., Speck C., Li H. (2021). The structure of ORC-Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6. Nat Commun 12, 3883. https://doi.org/10.1038/s41467-021-24199-1
Yuan Z., Schneider S., Dodd T., Riera A., Bai L., Yan C., Magdalou I., Ivanov I., Stillman B., Li H., and Speck C. (2020). Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6. Proc Natl Acad Sci U S A. 117(30):17747-17756. doi: 10.1073/pnas.2006231117
Noguchi Y., Yuan Z., Bai L., Schneider S., Zhao G., Stillman B., Speck C., Li H. (2017). The cryo-EM structure of Mcm2-7 on DNA suggests a new lagging-strand DNA extrusion model. Proc Natl Acad Sci U S A. 114(45):E9529-E9538. doi:10.1073/pnas.1712537114
Yuan Z., Riera A., Bai L., Sun J., Nandi S., Spanos C., Chen ZA., Barbon M., Rappsilber J., Stillman B., Speck C. and Li H. (2017). Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol., 24, 316–324. https://doi.org/10.1038/nsmb.3372