“How do cells forget how to become different cell types over time? Can we make them remember again? ”
Every organism starts as one cell that has the potential to become every cell type in the adult body. However, as this cell begins to divide to create a complex organism, the daughter cells slowly lose this ability as their “fate” and final destination in the developing body is decided. At the molecular level, decisions made by the cells as they divide and specialise are enshrined by the molecules that influence activity of genes in their DNA.
These molecules and the influences they have on DNA are called epigenetic modifications: we want to find how these modifications are maintained during development and across the life course of an adult organism; and also how these modifications can be removed to restore the original potential of those early embryonic cells. Interestingly, this is something that happens during normal embryonic development, when the embryo starts to make its own germ cells- precursors of the sperm and the egg. Studying this process will thus help us not only to treat infertility, but importantly to understand the barriers to efficient regeneration and the pathologies that are linked to cells losing the memory of what they were supposed to do (such as seen in cancer).
“Understanding of maintenance and erasure of epigenetic memory will help us to manipulate cell fate in vivo and in vitro”
Development of any organism starts with a totipotent cell (zygote). Through series of cell divisions and differentiation processes this cell will give rise to the whole organism containing hundreds of specialised cells. While the cells at the onset of development have the capacity to generate all cell types (ie are toti-or pluripotent), this developmental capacity is progressively lost as the cells undertake cell fate decisions. At the molecular level, the memory of these events is laid down in a complex layer of epigenetic modifications at both the DNA and the chromatin level. This is known as the epigenetic memory.
The main research focus of our laboratory is trying to understand molecular processes that underlie global erasure of epigenetic information as well as the maintenance of this information across long periods of time in terminally differentiated postmitotic cells.
We focus on mouse development and the germline cycle as our experimental models and study the epigenetic reprogramming events that occur naturally in vivo during mouse development (in the mouse zygote and in the early mouse germ cells). We are particularly interested in:
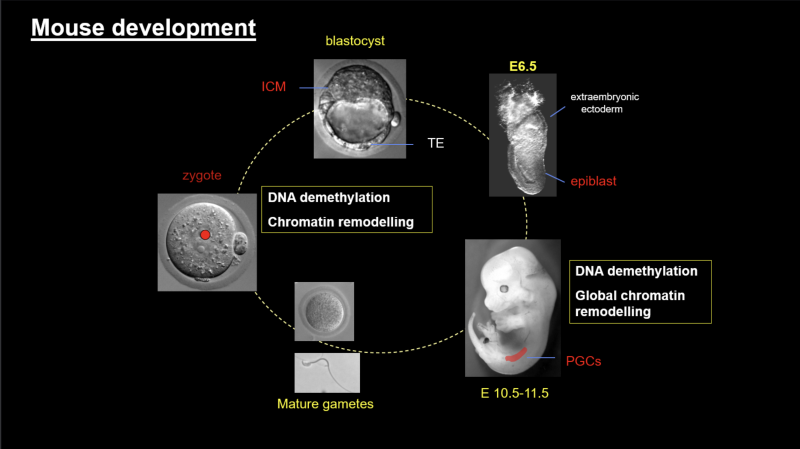
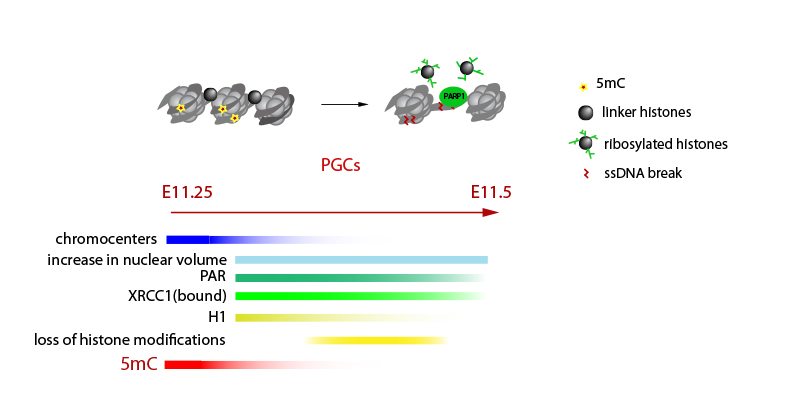
We are interested in the flow of epigenetic information across the germline cycle and between the generations, and how this can be influenced by the environment.
We investigate how DNA and histone modifications change when new germ cells are formed in the developing embryo and what changes are necessary for the nascent germ cells to undergo meiosis and to produce mature gametes. We are particularly keen to understand the development and long-term stability of the epigenetic information in the oocytes, the mechanistic implications for the age-related rapid decline in the oocyte quality and how this can be influenced by the environment. Our work has direct implications for our understanding of infertility and female reproductive potential.
Epigenetic information (DNA and histone modifications) is responsive to environmental stimuli. As a parallel interest of my team, we are looking at the mechanistic underpinning of the cross-talk between metabolism and epigenetic regulation. This has direct implications for our understanding of epigenetic changes observed in disease and during ageing.


Stewart-Morgan KR, Requena CE, Flury V, Du Q, Heckhausen Z, Hajkova P, Groth A et al., 2023, Quantifying propagation of DNA methylation and hydroxymethylation with iDEMS, Nature Cell Biology, ISSN: 1465-7392
Huang T-C, Wang Y-F, Vazquez-Ferrer E, Theofel I, Requena CE, Hanna CW, Kelsey G, Hajkova P et al., 2021, Sex-specific chromatin remodelling safeguards transcription in germ cells, Nature, Vol: 600, Pages: 737-742, ISSN: 0028-0836
Luo C, Hajkova P, Ecker JR. (2018). Dynamic DNA methylation: In the right place at the right time. Science, 361, 1336-1340.
Leitch HG, Hajkova P. (2018). Eggs sense high-fat diet. Nature Genetics 50(3), 318-319.
Hill PWS, Leitch HG, Requena CE, Sun Z, Amouroux R, Roman-Trufero M, Borkowska M, Terragni J, Vaisvila R, Linnett S, Bagci H, Dharmalingham G, Haberle V, Lenhard B, Zheng Y, Pradhan S, Hajkova P. (2018). Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 555, 392.
Rosic S, Amouroux R, Requena CE, Gomes A, Emperle M, Beltran T, Rane JK, Linnett S, Selkirk ME, Schiffer PH, Bancroft AJ, Grencis RK, Jeltsch A, Hajkova P, Sarkies P. (2018). Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nature Genetics 50, 452–459.
Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D’Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P. (2016). De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 18, 225.
Nashun B, Hill PW, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, Kelsey G, Hajkova P. (2015). Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Molecular Cell 60(4), 611-25.
Nashun B, Hill PW, Hajkova P. (2015). Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO Journal 34(10), 1296-308. Pii: e201490649.
Hill PW, Amouroux R, Hajkova P. (2014). DNA demethylation, tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story. Genomics 104, 324-333.
Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nature Structural & Molecular Biology 20(3), 311–316.
Hajkova P. (2011). Epigenetic reprogramming in the germline: towards the ground state of the epigenome. Philosophical Transactions of the Royal Society B 366(1575), 2266–2273.
Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. (2010). Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329(5987), 78–82.
Hajkova P. (2010). Epigenetic reprogramming – taking a lesson from the embryo. Current Opinion in Cell Biology 22(3), 342–350.
Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. (2008). Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452(7189), 877–881.
Surani MA, Hayashi K, Hajkova P. (2007). Genetic and epigenetic regulators of pluripotency. Cell 128(4), 747–762.