We create tools that help us understand biology and design organisms with new traits.
Creating molecular technologies
We create molecular tools and technologies to address specific, intriguing challenges at the intersection of protein engineering and synthetic biology. We currently focus on two main directions:
— Protein design for therapeutics and crop protection,
— Self-sustained luminescence as a reporter tool for agricultural and biomedical applications.
What does AI-based protein design enable in therapeutics and agriculture?
Evolutionary pressures equipped organisms with an amazing diversity of protein-based defence and attack mechanisms. A number of these have been adopted as therapeutic or crop protection molecules. Developments in de novo protein designs promise great expansion of arsenal of such proteins. We focus on solving the downstream bottlenecks in protein design pipelines and creating a robust platform for their development.
Can we use self-sustained luminescence to visualise molecular events at the organism scale?
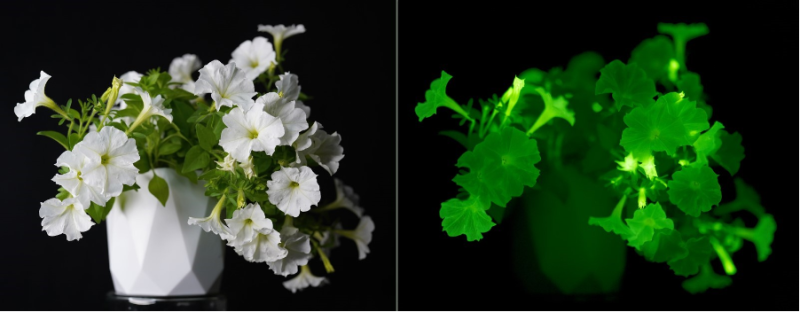
Creating organisms that glow in the dark requires understanding of biochemical pathways that lead to bioluminescence. In 2018, our collaborators and we discovered a mechanism of light emission in fungi, where bioluminescence results from activity of “caffeic acid cycle” – a short metabolic pathway catalysed by four enzymes. We showed that integration of that pathway into host metabolism allows engineering of organisms with self-sustained luminescence.
We are now working on applying self-sustained luminescence to visualise invisible physiological events. By making expression of bioluminescence genes dependent on physiological or environmental events, we can monitor virtually any molecular event completely non-invasively. For example, luminescence can show hormone activity in tissues, patterns of gene expression, or be used to create sentinel organisms that report otherwise invisible environmental cues.
For more information about the lab, visit our website: https://designing.bio
“We create molecular technologies to study life and to engineer organisms with new behaviours”
We work in several research directions, both basic and applied, from studying structure-function relationship in proteins to engineering bacterial strains that can perform directed evolution without human involvement. We combine our interest in new technologies, high-throughput assays and automation to engineer organisms.
Previously, we have worked on fluorescent protein development, high-through put assays for measuring protein activities, mechanisms of bioluminescence and development of transgenic bacteria and plants.
At the MRC LMS we are approaching these and other problems from the perspective of synthetic biology, investing time into automation of our molecular biology routines and creating new molecular technologies for light-based computation and communication of cells. Additionally, we will work on studying structure-function relationship in proteins with the aim to compare properties of various regions of the protein sequence space in a systematic manner.
For more information visit the lab website: https://designing.bio.
Supplementary video 1 from Sarkisyan KS*, Bolotin DA*, Meer MV, et al. (2016). Local fitness landscape of the green fluorescent protein. Nature 533, 397–401. doi:10.1038/nature17995. This is the 3D-rendering of our dataset that is also depicted in Figure 1b in the article. The protein sequence is arranged in a circle, with the N terminal and the chromophore labelled on the outer circle. Black line markers outside the fitness landscape representation are positioned every 10 sites of avGFP. The Z-axis, height, represents the level of fluorescence, which is colour-coded from green to black. The surface is shown as the median fluorescence brightness levels of all mutations at a given site with fluorescence levels conferred by individual mutations shown by dots. The centre represents the fluorescence of avGFP with distance away from it corresponding to the number of mutations in the genotype. The median surface extends up to genotypes with 10 mutations.
Our group focuses on developing new technologies to program cellular processes. We use protein engineering, modelling, and synthetic biology to create genetic circuits that predictably change gene expression and metabolism.
In the past, our work focused on systematic understanding of mutational effects in proteins through high-throughput genotype-phenotype mapping, and on creating imaging technologies based on self-sustained luminescence.
Our current focus is on designing molecular building blocks to engineer new traits in organisms. Our main models are bacterial and plant cells, which we use for prototyping genetic circuits, and for production of novel compounds with potential clinical applications. We then combine them with animal models to test effects on animal physiology and behaviour.

Shakhova, E. S., Karataeva, T. A., Markina, N. M., Mitiouchkina, T., Palkina, K. A., Perfilov, M. M., Wood, M. G., Hoang, T. T., Hall, M. P., Fakhranurova, L. I., Alekberova, A. E., Malyshevskaia, A. K., Gorbachev, D. A., Bugaeva, E. N., Pletneva, L. K., Babenko, V. V., Boldyreva, D. I., Gorokhovatsky, A. Y., Balakireva, A. V., . . . Mishin, A. S. (2024). An improved pathway for autonomous bioluminescence imaging in eukaryotes. Nature Methods. https://doi.org/10.1038/s41592-023-02152-y
Palkina, K. A., Karataeva, T. A., Perfilov, M. M., Fakhranurova, L. I., Markina, N. M., Somermeyer, L. G., Garcia-Perez, E., Vazquez-Vilar, M., Rodriguez-Rodriguez, M., Vazquez-Vilriales, V., Shakhova, E. S., Mitiouchkina, T., Belozerova, O. A., Kovalchuk, S. I., Alekberova, A., Malyshevskaia, A. K., Bugaeva, E. N., Guglya, E. B., Balakireva, A., . . . Sarkisyan, K. S. (2024). A hybrid pathway for self-sustained luminescence. Science Advances, 10(10). https://doi.org/10.1126/sciadv.adk1992
Somermeyer, L. G., Fleiss, A., Mishin, A. S., Bozhanova, N. G., Igolkina, A. A., Meiler, J., Pujol, M. A., Putintseva, E. V., Sarkisyan, K. S., & Kondrashov, F. A. (2022). Heterogeneity of the GFP fitness landscape and data-driven protein design. eLife, 11. https://doi.org/10.7554/elife.75842
Mitiouchkina, T., Mishin, A. S., Somermeyer, L. G., Markina, N. M., Chepurnyh, T. V., Guglya, E. B., Karataeva, T. A., Palkina, K. A., Shakhova, E. S., Fakhranurova, L. I., Chekova, S. V., Tsarkova, A. S., Golubev, Y. V., Negrebetsky, V. V., Dolgushin, S. A., Shalaev, P. V., Shlykov, D., Melnik, O. A., Shipunova, V. O., . . . Sarkisyan, K. S. (2020). Plants with genetically encoded autoluminescence. Nature Biotechnology, 38(8), 944–946. https://doi.org/10.1038/s41587-020-0500-9
Ding, S. S., Romenskyy, M., Sarkisyan, K. S., & Brown, A. E. X. (2020). Measuring Caenorhabditis elegans Spatial Foraging and Food Intake Using Bioluminescent Bacteria. Genetics, 214(3), 577–587. https://doi.org/10.1534/genetics.119.302804
Kotlobay, A. A., Sarkisyan, K. S., Mokrushina, Y. A., Marcet-Houben, M., Serebrovskaya, E. O., Markina, N. M., Somermeyer, L. G., Gorokhovatsky, A. Y., Vvedensky, A., Purtov, K. V., Petushkov, V. N., Rodionova, N. S., Chepurnyh, T. V., Fakhranurova, L. I., Guglya, E. B., Ziganshin, R., Tsarkova, A. S., Kaskova, Z. M., Shender, V., . . . Yampolsky, I. V. (2018). Genetically encodable bioluminescent system from fungi. Proceedings of the National Academy of Sciences, 115(50), 12728–12732. https://doi.org/10.1073/pnas.1803615115
Sarkisyan, K. S., Bolotin, D. A., Meer, M. V., Usmanova, D. R., Mishin, A. S., Sharonov, G. V., Ivankov, D. N., Bozhanova, N. G., Baranov, M. S., Soylemez, O., Bogatyreva, N. S., Vlasov, P. K., Egorov, E. S., Logacheva, M. D., Kondrashov, A. S., Chudakov, D. M., Putintseva, E. V., Mamedov, I. Z., Tawfik, D. S., . . . Kondrashov, F. A. (2016). Local fitness landscape of the green fluorescent protein. Nature, 533(7603), 397–401. https://doi.org/10.1038/nature17995