“What do cell membrane transporters tell us about drug uptake and disease?”
Membrane transporters are proteins that allow substances into or out of a cell. They act as molecular gatekeepers and regulate cellular uptake of nutrients and ions as well as the release of waste products.
Alterations in transporter expression levels are a vital part of cancer-specific metabolic changes, since they allow the tumour to efficiently scavenge nutrients from its micro-environment in order to maintain proliferation.
We now know that many drugs “hijack” these transporters to enter the cell, so cancerous cells may also have an altered sensitivity to certain treatments as a result. However, more understanding is needed.
We combine analysis of large-scale, publicly available datasets with cell biology, gene editing, and mass spectrometry (a method that detects and quantifies small molecules) to better identify the specific transporters involved in drug uptake.
We work to determine how these transporters affect the cells sensitivity to the drug, and how this impacts the toxicity of the compound in certain cell types and tissues.
We hope that with a better understanding of the role of transporters in drug uptake we can try to exploit these findings to increase the efficiency of drug delivery. This could help transform cancer treatments, and even pave the way for tailored treatment strategies based on the transporter expression of a patient’s tumour.
It could also help predict the efficacy of drugs in clinical trials, and predict potentially toxic effects of drugs in normal tissues.
“Our research aims to understand how membrane transporters mediate drug uptake and how this intersects with their role in cancer metabolism.”
Membrane transporters act as molecular gatekeepers to the cell by regulating the uptake of nutrients and ions as well as the release of waste products. Alterations in transporter expression levels are a vital part of cancer-specific metabolic changes, since they allow the tumour to efficiently scavenge nutrients from its micro-environment in order to maintain proliferation.
It is becoming increasingly clear that a large number of drugs ‘hijack’ plasma membrane transporters in order to enter the cell, rather than passing through the lipid bilayer. This means that as well as influencing cancer metabolism, the differential expression of transporters in cancer could define the drug sensitivity profile of a tumour. To date, however, the transporters required for cell uptake have only been identified for a handful of drugs, and the lack of understanding of drug entry into cells could partially explain the failure of many compounds in clinical trials.
We combine analysis of large-scale, publicly available datasets with cell biology, CRISPR gene-editing and metabolomics to identify the specific transporters involved in drug uptake, and to determine how expression of these transporters affects specificity, selectivity and ultimately toxicity of the compound. By also studying the role of the transporter in cancer metabolism, we can try to exploit its underlying biology to drive efficient drug uptake.
Identification of transporter-drug interactions could provide functional biomarkers to enable tailoring of treatment strategy based on a patient’s personal transporter expression patterns. More broadly, this knowledge could help to predict efficacy of drugs in the clinic, to reduce the failure rate of new compounds in clinical trials, and could also be used to predict off-target and potentially toxic effects of drugs in normal tissues.
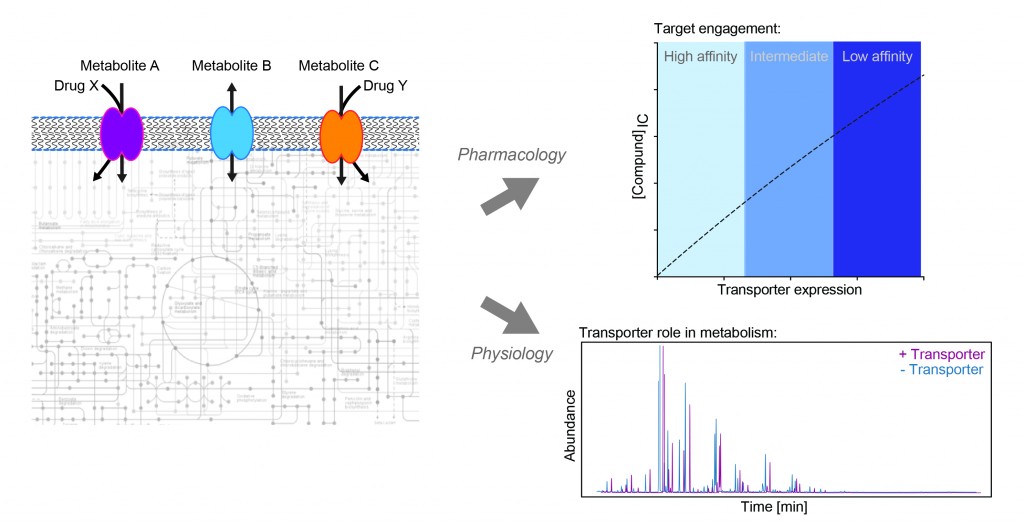
Our research aims to understand the role of transporter proteins in cancer. This class of membrane proteins can influence drug response in two ways: either directly, by transporting a drug into or out of a cell; or indirectly, by shaping the metabolic state of the cell, which in turn alters the efficacy of that therapy. Transporter expression in cancer is highly heterogeneous, so by better understanding of which transporters influence drug activity and how they do this, we may be able to better predict drug response and design more personalised treatment strategies for patients.
While we are interested in a range of therapeutic drugs and cancer types, to date, our work mainly focuses on ovarian and lung cancer.



Moncayo, C. R., Restuadi, R., Zhang, G., Marks, D., Ortega-Prieto, P., Doherty, E., Lambie, N., Whilding, C., Andrew, I., Montoya, A., Patel, B., Pennycook, B. R., Wu, V., Takats, Z., Matthews, N., Young, G. R., Shliaha, P., Game, L., Lenhard, B., McNeish, I., Fotopoulou, C., Barr, A. R., Cunnea, P., Hall, Z., & Fets, L. (2025). Multimodal imaging reveals a lysosomal drug reservoir that drives heterogeneous distribution of PARP inhibitors [Preprint]. bioRxiv https://doi.org/10.1101/2025.05.31.656628
Alam, S., Doherty, E., Ortega-Prieto, P., Arizanova, J., & Fets, L. (2023). Membrane transporters in cell physiology, cancer metabolism and drug response. Disease Models & Mechanisms 16(11). https://doi.org/10.1242/dmm.050404
Fets, L., Bevan, N., Nunes, P.M., Campos S., Silva Dos Santos M., Sherriff E., MacRae J. I, House D., Anastasiou D. (2022). MOG analogues to explore the MCT2 pharmacophore, α-ketoglutarate biology and cellular effects of N-oxalylglycine. Commun Biol 5, 877. https://doi.org/10.1038/s42003-022-03805-y
Newton H, Wang Y, Camplese L, Mokochinski JB, Kramer HB, Brown AEX, Fets L, Hirabayashi S. (2020). Systemic muscle wasting and coordinated tumour response drive tumourigenesis. Nature Communications 11, 4653.
Fets L, Driscoll PC, Grimm F, Jain A, Nunes PM, Gounis M, Doglioni G, Papageorgiou G, Ragan TJ, Campos S, dos Santos MS, MacRae JI, O’Reilly N, Wright AJ, Benes CH, Courtney KD, House D, Anastasiou D. (2018). MCT2 mediates concentration-dependent inhibition of glutamine metabolism by MOG. Nat Chem Biol 14, 1032–1042.
Macpherson JA, Theisen A, Masino L, Fets L, Driscoll PC, Encheva V, Snijders AP, Martin SR, Kleinjung J, Barran PE, Fraternali F, Anastasiou D. (2019). Functional cross-talk between allosteric effects of activating and inhibiting ligands underlies PKM2 regulation. eLife 2019;8:e45068.
Clark J, Kay RR, Kielkowska A, Niewczas I, Fets L, Oxley D, et al. Dictyostelium uses ether-linked inositol phospholipids for intracellular signalling. (2014). EMBO J 33(19), 2188–2200.
Fets L, Nichols JME, Kay RR. A PIP5 kinase essential for efficient chemotactic signaling. (2014). Curr Biol 24(4), 415–21.