To live healthy lives, we need each of our organs to do different jobs: The brain needs to think, feel, and direct our movements, the gut needs to digest food, the heart needs to pump blood and nutrients through our bodies. Even within a particular organ, there are many different types of cells, all with their very own roles. Red blood cells, for example, carry oxygen, and white blood cells fight infections.
The instructions for all these functions are laid down in our genome. And while our cells do very different jobs, the genome is the same in each and every cell. So how, you may wonder, can a brain cell ‘know’ to do brain things, a liver cell to do liver things, and so on for all of the hundreds of cell types that make up a human?
These are exactly the questions we try to answer with our research: How does each cell know what type of cell it is, and therefore what parts of the genome it needs to read for instructions how to do its job? What goes wrong when this order breaks down, resulting in disease, and what can be done to get it fixed?
Please click on the “Our Science: The Detail” tab above to read more.
“We explore gene regulatory mechanisms that drive mammalian cell type specification.”
We explore how gene regulatory mechanisms drive cell type specification with the aim of addressing important challenges in biomedical research:
1) How are cell fates established by the interplay of transcription factors, epigenetic regulators, and chromatin state?

2) How is the genome organised in the 3D space of the nucleus to enable genome function?
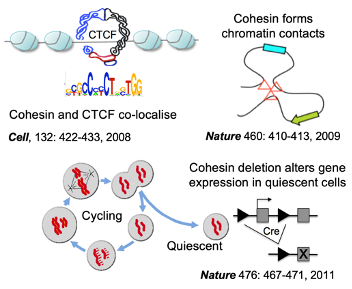
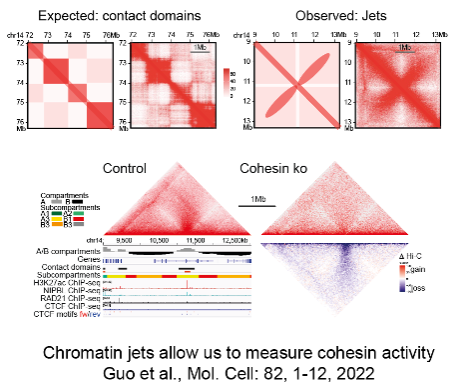
3D gene control is a fundamental biological mechanism, and relevant to understanding the impact of cohesin mutations on human development and cancer.
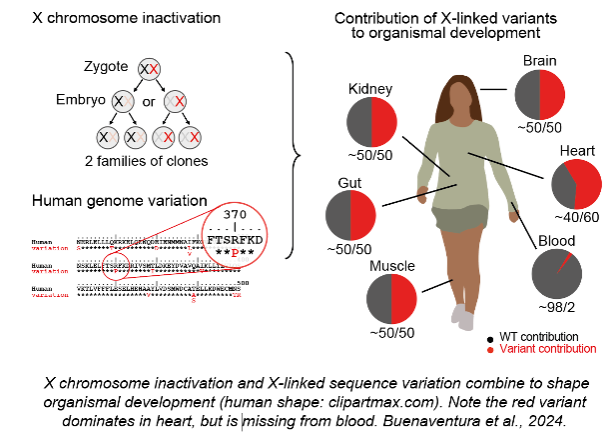
3) How does X-linked sequence variation shape organismal development?
Towards these aims, we collaborate widely and embrace working across disciplines.
We combine genetics, cell biology, computational biology, maths & medicine.
Our approach drives innovation and assures interdisciplinary training for our Masters- and PhD students, post-docs and technicians.
Our work promotes transdisciplinary scientific research and training.
We discover mechanisms of human development and disease to find new therapeutic targets and use preclinical models to scope the potential for improving existing conditions.
We explore how genetic variation impacts disease susceptibility, progression and reversal.
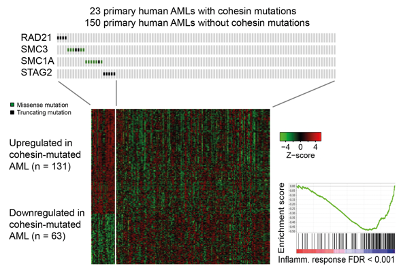
Cohesin is a protein complex that holds sister chromatids together; it facilitates genomic functions ranging from gene expression to genome replication and repair, and is mutated in multiple cancer types, including acute myeloid leukaemia (AML). We found that cohesin mutations confer a selective advantage to leukemic stem cells by dampening inflammation and increasing self-renewal. Of relevance to patient stratification, mutation type can affect drug efficacy. Inherited cohesin mutations cause Cornelia de Lange Syndrome. We investigate whether correcting mutations can reverse disease in pre-clinical model.
The IKZF1 gene encodes a gene regulatory protein and is recurrently mutated in acute B cell progenitor leukaemia (B-ALL). Towards new treatments for B-ALL we investigate the impact of IKZF1 mutations in B-ALL.
A new direction of research is the sex-specific impact of human genetic variation with implications for diagnosis and treatment of X-linked diseases.


Buenaventura T, Bagci H, Patrascan I, Graham JJ, Hipwell KD, Oldenkamp R, King JWD, Urtasun J, Young G, Mouzo D, Gomez-Cabrero D, Rowland BD, Panne D, Fisher AG, Merkenschlager M. (2024). Competition shapes the landscape of X-chromosome-linked genetic diversity. Nature Genetics 56: 1678-88
Al-Jibury E, King JWD, Guo Y, Lenhard B, Fisher AG, Merkenschlager M, Rueckert D. (2023). A deep learning method for replicate-based analysis of chromosome conformation contacts using Siamese neural networks. Nature Communications 14: 5007. doi: 10.1038/s41467-023-40547-9.
Guo Y, Al-Jibury E, Garcia-Millan R, Ntagiantas K, King JWD, Nash AJ, Galjart N, Lenhard B, Ruecekert D, Fisher AG, Pruessner G, Merkenschlager M. (2022). Chromatin jets define properties of cohesin-driven in vivo loop extrusion. Molecular Cell 82, 1-12
Robles-Rebollo I, Cuartero S, Canellas A, Wells S, Karimi MM, Mereu E, Chivu AG, Heyn H, Whilding C, Dormann D, Marguerat S, Rioja I, Prinjha RK, Stumpf MPH, Fisher AG, Merkenschlager M. (2022). Cohesin couples transcriptional bursting probabilities of inducible enhancers and promoters. Nature Communications 13: 4342 https://doi.org/10.1038/s41467-022-31192-9
Dsouza KB, Maslova A, Al-Jibury E, Merkenschlager M, Bhargava VK, Libbrecht MW. (2022). Learning representations of chromatin contacts using a recurrent neural network identifies genomic drivers of conformation. Nature Communications 13: 3704 https://doi.org/10.1038/s41467-022-31337-w
Calderon L, Weiss FD, Beagan JA, Oliveira MA, Georgieva R, Wang YF, Carroll T, Dharmalingam G, Gong W, Tossell K, de Paola V, Whilding C, Ungless MA, Fisher AG, Phillips-Cremins JE, Merkenschlager M. (2022). Cohesin-dependence of neuronal gene expression relates to chromatin loop length. eLife 11: e76539 https://doi.org/10.7554/eLife.76539
Karimi MM, Guo Y, Cui X, Pallikonda HA, Horková V, Wang Y-F, Ruiz Gil S, Rodriguez-Esteban G, Robles-Rebollo I, Bruno L, Georgieva R, Patel B, Elliott J, Dore MH, Dauphars D, Krangel MS, Lenhard B, Heyn H, Fisher AG, Štěpánek O, Merkenschlager M. (2021). The order and logic of CD4 CD8 lineage choice and differentiation in mouse thymus. Nature Communications 12: 99. https://doi.org/10.1038/s41467-020-20306-w
Weiss FD, Calderon L, Wang YF, Georgieva R, Guo Y, Cvetesic N, Kaur M, Dharmalingam G, Krantz ID, Lenhard B, Fisher AG, Merkenschlager M. 2021. Neuronal genes deregulated in Cornelia de Lange Syndrome respond to removal and re-expression of cohesin. Nature Communications 12: 2919 https://doi.org/10.1038/s41467-021-23141-9
Djeghloul D, Patel B, Kramer H, Dimond A, Whilding C, Brown K, Kohler A, Feytout A, Veland N, Elliott J, Bharat TAM, Tarafder AK, Löwe J, Ng BL, Guo Y, Guy J, Huseyin MK, Klose RJ, Merkenschlager M, Fisher AG. (2020). Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction. Nature Communications, 11, 4118.
Van de Pette M, Galvao A, Millership SJ, To W, Dimond A, Prodani C, McNamara G, Bruno L, Sardini A, Webster Z, McGinty J, French P, Uren AG, Castillo-Fernandez J, John RM, Ferguson-Smith AC, Merkenschlager M, Kelsey G, Fisher AG. (2020). Epigenetic change induced by in utero dietary challenge provokes phenotypic variability across multiple generations of mice. bioRxiv, https://doi.org/10.1101/2020.08.07.241034
Bruno L, Ramlall V, Studer RA, Sauer S, Bradley D, Dharmalingam G, Carroll T, Ghoneim M, Chopin M, Nutt SL, Elderkin S, Rueda DS, Fisher AG, Siggers T, Beltrao P, Merkenschlager M. (2019). Selective deployment of transcription factor paralogs with submaximal strength facilitates gene regulation in the immune system. Nature Immunology, https://doi.org/10.1038/s41590-019-0471-5
Ferreirós-Vidal I, Carroll T, Zhang T, Lagani V, Ramirez RN, Ing-Simmons E, Gómez-Valadés AG, Cooper L, Liang Z, Papoutsoglou G, Dharmalingam G, Guo Y, Tarazona S, Fernandes SJ, Noori P, Silberberg G, Fisher AG, Tsamardinos I, Mortazavi A, Lenhard B, Conesa A, Tegner J, Merkenschlager M, Gomez-Cabrero D. (2019). Feedforward regulation of Myc coordinates lineage-specific with housekeeping gene expression during B cell progenitor cell differentiation. PLoS Biology, 12; 17(4).
Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Irene Robles-Rebollo, Xiao X, Wang Y-F, Barozzi I, Djeghloul D, Amano MT, Niskanen H, Petretto E, Dowell RD, Tachibana K, Kaikkonen MU, Nasmyth KA, Lenhard B, Natoli G, Fisher AG, Merkenschlager M. (2018). Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nature Immunology 19, 932–941.
Liang Z, Brown KE, Carroll T, Taylor B, Ferreiros Vidal I, Hendrich B, Rueda D, Fisher AG, Merkenschlager M. (2017). A high-resolution map of transcriptional repression. eLife 6, ISSN:2050-084X. DOI: 10.7554/eLife.22767.
Van de Pette M, Abbas A, Feytout A, McNamara G, Bruno L, To WK, Dimond A, Sardini A, Webster Z, McGinty J, Paul EJ, Ungless MA, French PMW, Withers DJ, Uren A, Ferguson-Smith AC, Merkenschlager M, John RM, Fisher AG. (2017). Visualizing Changes in Cdkn1c Expression Links Early-Life Adversity to Imprint Mis-regulation in Adults. Cell Reports 18(5):1090-1099. DOI: http://dx.doi.org/10.1016/j.celrep.2017.01.010.
Fisher AG, Stumpf MPH, Merkenschlager M. (2017). Reconciling Epigenetic Memory and Transcriptional Responsiveness. Cell Systems 4(4):373-374. doi: 10.1016/j.cels.2017.04.005. PMID: 28448796.
Merkenschlager M, Nora EP. (2016). CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annual Review of Genomics and Human Genetics 17: 17-43, ISSN: 1527-8204.
Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, Fisher AG, Lenhard B, Merkenschlager M. (2015). Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Research 25 (4), 504-513. ISSN: 1088-9051.
Lavagnolli T, Gupta P, Hörmanseder E, Mira-Bontenbal H, Dharmalingam G, Carroll T, Gurdon JB, Fisher AG, Merkenschlager M. (2015). Initiation and maintenance of pluripotency gene expression in the absence of cohesin. Genes & Development 29, 23-38. ISSN: 0890-9369.
Blevins R, Bruno L, Carroll T, Elliott J, Marcais A, Loh C, Hertweck A, Krek A, Rajewsky N, Chen CZ, Fisher AG, Merkenschlager M. (2015). microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes. PLOS Genetics 11. ISSN: 1553-7390.
Marcais A, Blevins R, Graumann J, Feytout A, Dharmalingam G, Carroll T, Amado IF, Bruno L, Lee K, Walzer T, Mann M, Freitas AA, Boothby M, Fisher AG, Merkenschlager M. (2014). microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. Journal of Experimental Medicine 211, 2281-2295. ISSN: 0022-1007.
Merkenschlager M, Odom DT. (2013). CTCF and Cohesin: Linking Gene Regulatory Elements with Their Targets Cell 152 (6), 1285-1297. ISSN: 0092-8674.
Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, Flicek P, Dekker J, Merkenschlager M. (2013). Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Research 23 (12), 2066–2077.
Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, Kriaucionis S, Dawlaty MM, Jaenisch R, Merkenschlager M, Fisher AG. (2013). Different roles for tet1 and tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Molecular Cell 49 (6), 1023–1033.
Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. (2013). DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell 152 (4), 873–883.
Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. (2011). A role for cohesin in t-cell-receptor rearrangement and thymocyte differentiation. Nature 476 (7361), 467–471.
Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. (2009). Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460 (7253), 410–413.
Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132 (3), 422–433.