We aim to understand the epigenetic mechanisms that influence which genes will be switched ‘on’ and ‘off’ and how this is maintained. Ultimately, we investigate how cells know who are, and what this means for the development of diseases like Friedreich’s ataxia, rheumatoid arthritis, and many more.
“How do cells know who they are and what happens when things go wrong?”
There are around 200 different cell types in the human body. To code for the correct cell type, individual cells need to decide which genes to turn on and off, and remember these decisions every time it divides into daughter cells.
The process of turning these genes on and off and remembering in which way to do so is controlled by molecular regulators in the cells called epigenetic mechanisms.
Our research looks to better understand the causes of Friedreich’s ataxia (FA), a neurological disease that is caused by a faulty specific epigenetic mechanism. We are also studying how epigenetic mechanisms differ between the sexes.
For our genetic studies, we use genomic sequencing processes.
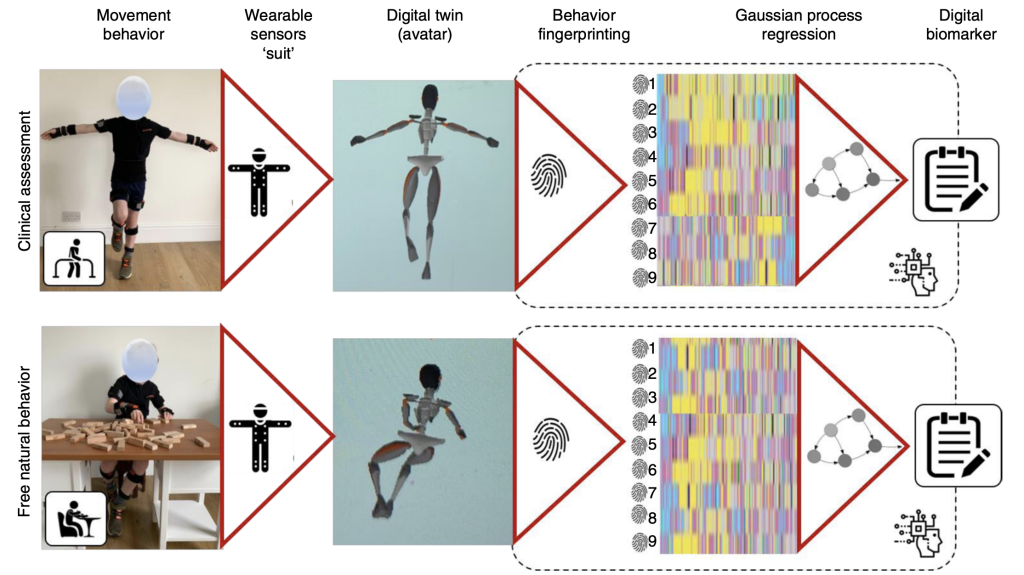
For FA research, we use a combination of clinical trials using patients and genomic sequencing. In partnership with researchers at UCL and UCLH, we are developing new motion-capture technology combined with AI and machine learning to monitor the effectiveness of treatments of this disease.
There are several diseases that effect people differently depending on their sex – this includes severe COVID and other infections, as well as autoimmune diseases like Rheumatoid Arthritis, Systemic Lupus Erythematosus (SLE) and Multiple Sclerosis (MS).
“We investigate how cells know who they are”
Around 200 types of cells make up the human body, each with its own unique shape and function. To code for the correct cell type, individual cells need to decide which genes they should switch ‘on’ or ‘off’ during development. Importantly, each cell must ‘remember’ this decision every time it divides. The process and maintenance of switching genes ‘on’ and ‘off’ is central to the biology of a multicellular organism and is influenced by epigenetic mechanisms.
The Epigenetic Mechanisms and Disease research group aims to understand the regulators, also known as epigenetic mechanisms, that influence which genes will be switched ‘on’ and ‘off’ and how this is maintained. Their studies on the differences between the sexes is regulated at an epigenetic level with a key epigenetic regulator being key – this finding is likely to be important for understanding sex-bias in diseases, for example susceptibility to severe COVID and other infections as well as autoimmune diseases such as Rheumatoid Arthritis, SLE and MS (Wijchers et al., 2010).
This group is led by Professor Richard Festenstein, a Clinical Professor of Molecular Medicine specialising in neurogenetics. His work on gene-switching led to the discovery that repeating units of DNA that occur in neurological diseases can switch ‘off’ genes leading to specific diseases. Richard has done a large amount of work on the neurological disease Friedreich’s ataxia (FA), a disease also caused by these repeating units of DNA. Friedreich’s ataxia is a progressive neurological disease causing a decline in balance and coordination as well as causing heart disease. Professor Festenstein showed that the FA gene known as Frataxin is switched ‘off’ by an epigenetic mechanism and can be switched on again by an epigenetic modifier
This research, contrary to previous beliefs, showed that heterochromatin dynamically allows DNA to be accessed (Science, 2003).
FA is a rare disease affecting 1/50 000 in the UK, making it difficult to develop treatments as a large number of patients are needed for each clinical trial. Richard’s research alongside Professor Faisal’s team devised a novel way of monitoring the effectiveness of treatments for this disease (Nature Medicine, 2023) using motion-capture technology combined with AI and machine learning. Their research continued with colleagues at UCL and UCLH to show that this technology also works in Duchenne Muscular Dystrophy (Nature Medicine, 2023). The findings of this research promise to make drug development for rare movement diseases more cheaply and quickly.
BBC 5 Live Interview with Professor Richard Festenstein and Yantia, a person with FA.
Explore the future of healthcare in our BBC Breakfast interview featuring a breakthrough AI technology. This AI analyses human movement more accurately and quickly than traditional methods, promising faster, cost-effective drug development. Watch to see how it’s transforming medical diagnostics.
Richard holds a fellowship with the Association of Physicians and the Royal College of Physicians
External Positions:





Kadirvelu, B., Gavriel, C., Nageshwaran, S., Ping Kei Chan J., Athanasopoulos, S., Ricotti, V., Voit, T., Giuniti, P., Festenstein, R & Faisal, A,. 2023 A wearable motion capture suit and machine learning predict disease progression in Friedreich’s ataxia. Nat Med, Vol:29, 86–94 (2023). ISSN 1546-170X
Ricotti, V., Kadirvelu, B., Selby, V., Festenstein, R ., Mercuri, E., Voit, T. & A. Faisal, A.,2023. Wearable full-body motion tracking of activities of daily living predicts disease trajectory in Duchenne muscular dystrophy. Nat Med, Vol:29, 95–103 (2023). ISSN: 1546-170X
Schon, KR., Horvath, R., Wei, W., Calabrese, C., Tucci,A., Ibanez,K., Ratnaike,T., Pitceathly,RDS.,
Bugiardini,E., Quinlivan,R., Hanna,MG., Clement,E., Ashton,E., Sayer,JA., Brennan,P.,
Josifova,D., Izatt,L., Fratter,C., Nesbitt,V., Barrett,T., McMullen,DJ.,Smith,A., Deshpande,C.,Smithson,SF., Festenstein,R., Canham,N., Caulfield,M., Houlden, H& Rahman, S,. 2021, Use of whole genome sequencing to determine genetic basis of suspected mitochondrial disorders: cohort study, British Medical Journal, Vol:375, ISSN:0959-535X
Reetz, K., Hilgers, R-D., Isfort, S., Dohmen, M., Didszun, C., Fedosov, K., Kistermann, J., Mariotti, C., Durr, A., Boesch, S., Klopstock, T., Rodríguez de Rivera Garrido, FJ., Schöls, L., Klockgether, T., Pandolfo, M., Korinthenberg, R., Lavin, P., Molenberghs, G., Libri, V., Giunti, P., Festenstein, R., Schulz, JB., 2019, Protocol of a randomized, double-blind, placebo-controlled, parallel-group, multicentre study of the efficacy and safety of nicotinamide in patients with Friedreich ataxia (NICOFA), Neurological Research and Practice, Vol:1, ISSN:2524-3489
Libri, V., Yandim C., Athanasopoulos S., Loyse, N., Natisvili, T., Pik Law, P., Kei Chan, P., Mohammad, T., Mauri, M., Tung Tam, K., Leiper, J., Piper, S., Ramesh, A., Parkinson, M., Huson, L., Giunti, P., Festenstein, R., 2014, Epigenetic and neurological effects and safety of high-dose nicotinamide in patients with Friedreich’s ataxia: an exploratory, open-label, dose-escalation study, The Lancet, Vol:384, ISSN:0140-6736, Pages:504-513
Chan, PK., Torres, R., Yandim, C., Law, PP., Khadayate, S., Mauri, M., Grosan, C., Chapman-Rothe, N., Giunti, P., Pook, M., Chan, PK., Torres, R., Yandim, C., Law, P., Khadayate, S., Mauri, M., Grosan, C., Chapman-Rothe, N., Giunti, P., Pook, M & Festenstein, R., 2013, Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreichs ataxia can be reduced upon HDAC inhibition by vitamin B3, Human Molecular Genetics, Vol:22, ISSN:0964-6906, Pages:2662-2675
Guillemette, B., Drogaris, P., Lin, H-HS., Armstrong, H., Hiragami-Hamada, K., Imhof, A., Bonneil, E., Thibault, P., Verreault, A., Festenstein, R., 2011, H3 lysine 4 8 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation, PLOS Genetics, Vol:7, ISSN:1553-7390, Pages:1-16
Wijchers PJ, Yandim C, Panousopoulou E, Ahmad, M; Harker, N; Saveliev, A; Burgoyne, PS & Festenstein, R., 2010, Sexual Dimorphism in Mammalian Autosomal Gene Regulation Is Determined Not Only by Sry but by Sex Chromosome Complement As Well, Developmental Cell, Vol:19, ISSN:1534-5807, Pages:477-484
Festenstein R, 2006, Breaking the silence in Friedreich’s ataxia, Nature Chemical Biology, Vol:2, ISSN:1552-4450, Pages:512-513
Saveliev, A., Everett C., Sharpe T., Webster, Z & Festenstein, R., 2003, DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing, Nature, Vol:422, ISSN:0028-0836, Pages:909-913
Festenstein, R., Pagakis, SN., Hiragami, K., Lyon, D., Verreault, A., Sekkali, B & Kioussis, D., 2003, Modulation of heterochromatin protein 1 dynamics in primary mammalian cells, Science, Vol:299, ISSN:0036-8075, Pages:719-721
Festenstein, R., Tolaini, M., Corbella, P., Mamalaki, C., Parrington, J., Fox, M., Miliou, A., Jones, M & Kioussis, D., 1996, Locus control region function and heterochromatin-induced position effect variegation, Science, Vol:271, ISSN:0036-8075, Pages:1123-1125