We strive to better understand the role that commensal microbes play in metabolism, disease and ageing and in the efficacy of drugs aimed at treating disease.
“How do our gut microbes impact our health?”
Animals, including humans, live in close association with billions of microbes, with all parties benefitting from the arrangement – for example, the microbes that live in your digestive system that help break down food. This symbiotic relationship forms what we call a the holobiont – a complex and interconnected system of organisms that essentially functions as one metabolic unit.
Everything the host eats effects the microbiota, and vice versa, everything the microbiota does could impact the host. This not only includes food, but medicines too.
Studies have shown that our gut microbes can actively impact human health. Their activity has been linked to nutrition-related syndromes like obesity and type-2 diabetes, as well as certain aging processes.
Our work focusses on better understanding how these microbial communities influence human physiology, as well as how their behaviour can impact or alter drug uptake. Some of our work has included testing how microbes in the gut could impact the uptake of first-line treatments for colorectal cancer.
We combine classical and advanced microbial genetics and high-throughput genomic/ chemical approaches. We have developed a novel system that allows us to find out more about the interactions between drugs, diet, microbe and host and we use two well-understood genetic models to do so; the bacterium E. coli and the nematode C. elegans.
We have already identified how microbes can produce metabolites that influence drug action. On the one hand we identified a signalling pathway in microbes that can be activated by host targeted drugs to regulate host physiology through the production of metabolites. On the other hand, we have also identified metabolites produced by microbes that can synergise with drugs, acting as adjuvant therapies.
We aim to better understand the mechanics of the holobiont, how the holobiont is impacted by drugs and other medication, and how organisms in the host may influence drug uptake and delivery.
We hope our work will pave the way for the development of new microbiota-based strategies that could improve the effectiveness of treatments for a range of diseases and disorders.
“Our lab focuses on the molecular mechanisms underlying metabolic diseases and ageing. These morbidities have a profound impact, both medically and also social and economically.”
Animals typically live in close association with commensal and symbiotic microbes forming a single metabolic unit – the Holobiont. Recent studies have revealed that the status of gut microbiota can influence nutrition-related syndromes such as obesity and type-2 diabetes, and ageing. However, to-date we know very little about how such interactions are regulated.
We focus on understanding how these microbial communities determine human physiology, and if they can be targeted by drugs to improve human health and ageing. We developed a complementary pharmacological pipeline with the potential to unravel drug-diet-microbe-host interactions. For this purpose, we utilise two tractable genetic models: the bacterium E. coli and the nematode C. elegans, widely used for host-microbiome research.
We combine classical and advanced microbial genetics and high-throughput genomic/chemical approaches with targeted metabolomics at the holobiont level. This strategy will identify drugable mechanisms in bacteria (e.g. signalling/biochemical pathways) that alter microbial metabolite availability with the capacity to regulate host physiology.
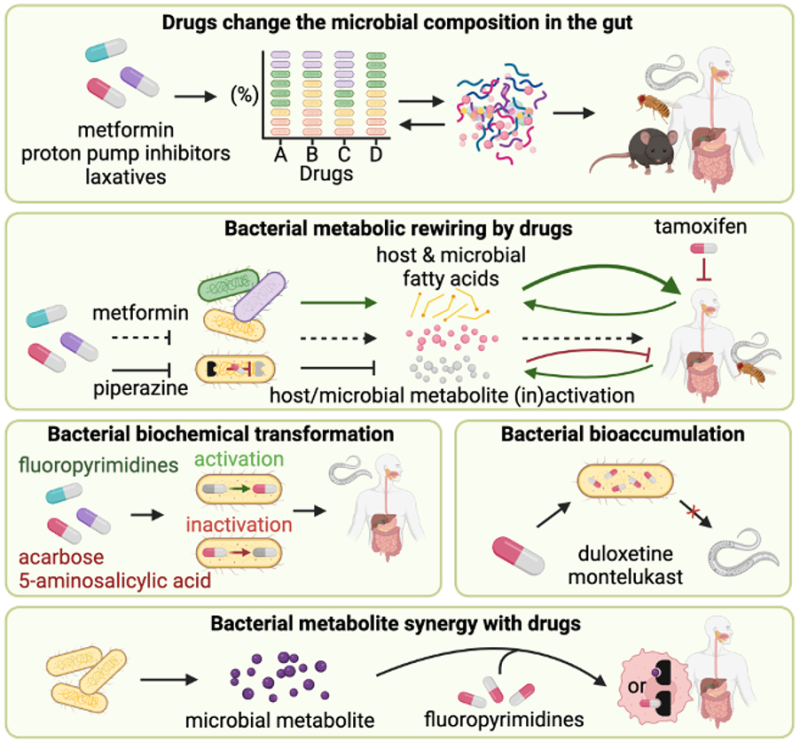
Therapeutic drugs are known to change gut microbial composition, with direct and indirect effects on microbial and host physiology. Conversely, microbes can alter drug efficacy directly by biochemical transformation or bioaccumulation of drugs, and indirectly via metabolites produced by the microbiota in a drug-induced dependent or independent alteration to microbial metabolism. Examples of model organisms used to reveal mechanisms at the interface of host-microbe-drug metabolism.




Essmann CL, Martinez-Martinez D, Pryor R, Leung K-Y, Krishnan KB, Lui PP, Greene NDE, Brown A, Pawar VM, Srinivasan MA, Cabreiro F. (2020). Mechanical properties measured by Atomic Force Microscopy define new health biomarkers in ageing C. elegans. Nature Communications 11 (1), 1-16.
Pryor R, Martinez-Martinez D, Quintaneiro L, and Cabreiro, F. (2020). The Role of the Microbiome in Drug Response. Annual Review of Pharmacology and Toxicology, 60, 417-435.
Rutter JW, Ozdemir T, Galimov ER, Quintaneiro LM, Rosa L, Thomas GM, Cabreiro F and Barnes CP. (2019). Detecting Changes in the Caenorhabditis elegans Intestinal Environment Using an Engineered Bacterial Biosensor. ACS Synthetic Biology 8 (12), 2620-2628.
Bana B and Cabreiro F. (2019). The Microbiome and Aging. Annu Rev Genet 53, 239-261
Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, De Haes W, Esser D, Waschina S, Lujan C, Smith RL, Scott TA, Martinez-Martinez D, Woodward O, Bryson K, Laudes M, Lieb W, Houtkooper RH, Franke A, Temmerman L, Bjedov I, Cochemé HM, Kaleta C, Cabreiro F. (2019). Host-Microbe-Drug-Nutrient screen identifies bacterial effectors of metformin therapy. Cell, 178(6), 1299-1312.
Norvaisas P and Cabreiro F. (2018). Pharmacology in the age of the holobiont. Current Opinion in Systems Biology, 10, 34-42.
Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY, Herrera-Dominguez L, Sudiwala S, Pessia A, Clayton PT, Bryson K, Velagapudi V, Mills PB, Typas A, Greene NDE, Cabreiro F. (2017). Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 169, 442-456 e418.
Cabreiro F. (2015). Metformin joins forces with microbes. Cell Host and Microbe, 19(1), 1-3.
Pryor R, Cabreiro F. (2015). Repurposing metformin: an old drug with new tricks in its binding pockets. Biochemical Journal, 471(3), 307-322.
Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cochemé HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. (2013). Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228-239.
Cabreiro F, Gems D. (2013). Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med 5, 1300-1310.
Coburn C, Allman E, Mahanti P, Benedetto A, Cabreiro F, Pincus Z, Matthijssens F, Araiz C, Mandel A, Vlachos M, Edwards SA, Fischer G, Davidson A, Pryor RE, Stevens A, Slack FJ, Tavernarakis N, Braeckman BP, Schroeder FC, Nehrke K, Gems D. (2013). Anthranilate fluorescence marks a calcium-propagated necrotic wave that promotes organismal death in C. elegans. PLoS Biol 11, e1001613.
Burnett C*, Valentini S*, Cabreiro F*, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. (2011). Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482-485.
* equally contributed.