By Sophie Arthur
February 25, 2019
Time to read: 5 minutes
CRISPR-Cas9 – the gene editing technology has gained worldwide recognition due to its huge potential across a range of sectors, but the gene editing tool also has its challenges and risks. New research from the MRC London Institute of Medical Sciences (LMS) may help to pave the way to improve the accuracy of gene editing for clinical applications.
Before we get excited by the potential applications in medicine, drug discovery and agriculture, we should all take a step back and actually question how this gene editing technology is working and understand what is really going on. That is exactly what the Single Molecule Imaging group at the LMS have looked into in collaboration with AstraZeneca.
The group led by David Rueda are interested in how the mechanism of Cas9 works and how this protein reaches any off-target effects to potentially improve the targeting and specificity of the CRISPR-Cas9 technology. In research published in the journal Nature Structural and Molecular Biology, the group reveal that Cas9 may be more promiscuous than people realise generating lots of off-target effects. A discovery that could have big implications on gene editing.
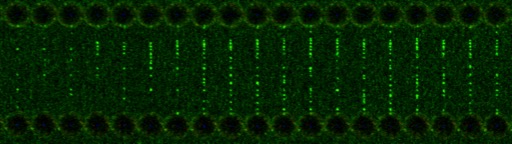
In this study, a single molecule of DNA was attached at either end to a bead and held by optical tweezers. The optical tweezers consist of those beads being trapped within a laser beam, thus holding the DNA in place. Those lasers can then be slowly moved apart bringing the bead with it and pulling the attached DNA. Think of a rubber band being pulled apart and stretched.
Using this ‘rubber band’ and optical tweezers, the researchers investigated how Cas9 reached its target by fluorescently labelling the protein and capturing those images using a new generation of powerful microscopes from LUMICKS to see where it is binding. The group revealed that when the DNA was in its relaxed position, that Cas9 bound specifically to its target sequence. However, as the structure of the DNA was distorted by stretching it apart, there was an influx of Cas9 binding to off target sites.
While this kind of pulling on the DNA is artificial, the distortion of the DNA structure occurs naturally and often in many of the general processes within a cell like gene expression, DNA repair and replication during cell division.
Cas9 is a type of enzyme called a nuclease; an enzyme that cuts a chain of nucleotides which includes DNA. Within the CRIPSPR-Cas9 gene editing technology, it is Cas9’s role to cleave the section of target DNA to allow for a gene to be repaired or even replaced with a healthy copy. After revealing that structural distortion of the DNA exposes off-target sites for Cas9 to bind to, it is important to work out whether these ‘molecular scissors’ are still functioning at these off-target sequences. So, the researchers at the LMS developed an assay to test whether Cas9 was binding and cleaving.
CASE PhD student Matt Newton, first author of the paper, discussed with us how DNA bubbles helped to verify whether Cas9 bound to off-target sites could still cut the DNA:
“The DNA bubbles come from the idea that stretching the DNA makes them easier to separate – something that was already hypothesised to be a key step in Cas9 binding. We generated stable DNA bubbles by putting bases in one strand that cannot pair with the other strand – essentially recreating a high force stretched piece of DNA. What we saw was that if you test the ability of Cas9 to bind the sites with as much as 50% mismatches, no binding is observed without a bubble but as soon as you generate a bubble and begin to separate the DNA strands, Cas9 can stably bind and cleave.”
While these assays show that Cas9 is unlikely to cut the DNA at these off-target sequences unless the DNA structure is distorted, it is important to remember that DNA is not a static molecule and is continually wound and unwound during fundamental cellular processes.
The results from this collaborative study between researchers at the LMS and AstraZeneca have highlighted the promiscuity of Cas9 in the current technology leading to more unintended gene edits than first thought.
But it is not all bad news as senior author David Rueda discusses:

“When we manipulated and stretched DNA structures, we found edits were made to unintended sites, which is a concern for using this gene-editing tool in lab and clinical settings. On the other hand, what’s very positive is we’ve developed a system that works to image CRISPR-Cas9 clearly so we can study why off-target sites are added and adapt the technology to improve on accuracy and specificity.”
Armed with the knowledge from this study, we can revolutionise the CRISPR-Cas9 gene editing technology as we know it. With a new generation of Cas9 that is more accurate, more specific and more efficient, we can take one step closer to it being used for therapeutic benefits.
‘DNA stretching induces Cas9 off-target binding and cleavage’ was published in Nature Structural and Molecular Biology on 25 February. Read the article here.