By Sophie Arthur
January 8, 2018
Time to read: 5 minutes
By Dr Sophie Arthur
It is not just the sequence of DNA that is crucial, but its structure too. DNA can form a variety of structures beyond its ‘traditional’ double-stranded helix from hairpins, to loops, to coils. Yet, each of those structures can have an impact on the cellular machinery and as a result their function.
There is another type of DNA structure called a G-quadruplex, or a quadruple helix, that is made up of four strands of DNA. It is becoming clearer that these structures can affect a number of biological processes too such as DNA replication, gene transcription and telomere maintenance, and are also found more frequently in cancer cells. These quadruplexes affect key processes that allow our DNA to be copied and read so genes can be switched on, and also play a role in various diseases. However, we don’t fully understand the consequences of these DNA structures because of a lack of tools and methods that allow researchers to study them in living cells.
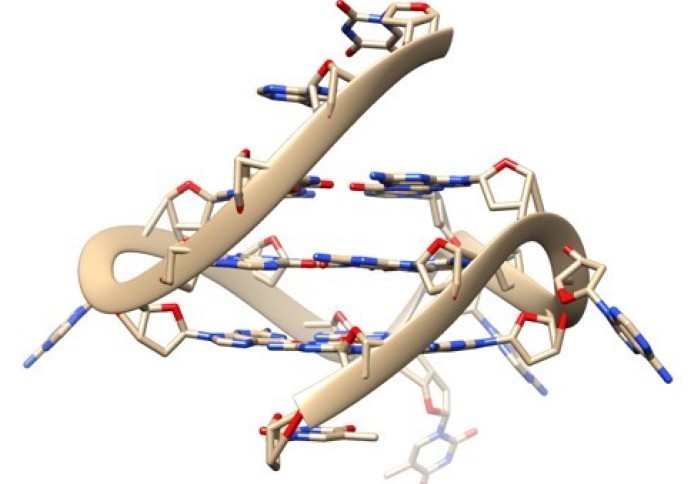
Research published today from the Telomere Stability and Replication group at the MRC LMS, in collaboration with scientists at Imperial College London, in Nature Communications reveals a new tool that allows researchers to examine these structures in living cells, and begin to decipher the effects of these quadruplexes on those cells. This collaboration between the LMS and the Department of Chemistry at Imperial was enabled by The President’s Excellence Fund for Frontier Science awarded in 2017.
This new tool takes the form of a fluorescent probe called DAOTA-M2, which glows or fluoresces when bound to quadruplexes. As there is far more double stranded DNA in a cell than the quadruplex DNA, it is “like finding a needle in a haystack, if that needle is also made of hay” as described by one of the lead authors Ben Lewis. As a result, you can’t make a probe selective enough. To overcome this issue, instead of analysing how bright the fluorescence is, the team measure how long the fluorescence lasts – a technique called fluorescence lifetime – to distinguish between the double helix and the quadruple helix DNA.
One of the benefits of this new technique is that now researchers can study the quadruplex structures in live cells. Previously, there were techniques that allowed researchers to study these DNA structures in so-called fixed cells, but that only allowed you to capture a snapshot in time. This new technique using living cells is like going from a photo to a video. Now, scientists can capture movements and effects across time, so you can examine what happened before or after that certain snapshot in time.
This probe can now be used in living cells to unravel the biology too. A helicase is an enzyme that can unwind these quadruplex structures in cells. This study is the first time that this method has been used to observe the effects of those helicases in living cells. The researchers found that if these helicases were removed from the cells, then more quadruplex DNA was present. This tells us that these helicases play a crucial role in removing these quadruplexes in cells. This had been shown indirectly previously, but this new probe has meant that the team have been able to directly show this in living cells.
Not only does this new technology allow researchers to study these structures more easily in the lab, but it could also help in the search for therapeutics. Some researchers are interested in G-quadruplex binding agents as potential drugs for diseases, such as cancer. The team have created a test where you can test a range of potential drug targets at once and see whether it displaces the fluorescent probe. In this test, the probe DAOTA-M2 is bound to quadruplexes in living cells. If a strong quadruplex binding agent is added to those cells and displaces the probe, the researchers will be able to see a change in the fluorescence lifetime, or the amount of time the fluorescence lasts. In this test, if researchers see that change, then it indicates that this binding agent is a good candidate for a therapeutic. Because this technology uses fluorescence, researchers can now see these interactions under the microscope. Something that would have been more difficult to see and understood previously.
Jean-Baptise Vannier, Head of the Telomere Stability and Replication group and one of the senior authors of this study, shared more about the study:
“This paper is a fruit of collaboration between my group at the LMS and also the Vilar and Kuimova groups at Imperial’s Department of Chemistry. It is a true interdisciplinary study and the lead authors Ben and Peter have been working on three fields of research to achieve this paper; chemistry, biology and optical physics. It is a tremendous achievement to be able to master specific techniques and technologies in three completely different disciplines to create a fantastic outcome. It puts us in a strong position in the field to study these molecules now.
In terms of next steps for this research, we are looking at ways to optimise and improve this tool so that we can collect data much more easily. But also, we want to use the tool to unravel the biology of these quadruplexes in living cells further and explore new biological problems.”
‘Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy’ was published on 8 January in the journal Nature Communications. Read the full article here.