By Sofia Velasquez-Pimentel
January 12, 2023
Time to read: 5 minutes
A new quantitative approach unravels complex dynamics of copying epigenetic information across cell generations
A new mass spectrometry-based approach that sheds light on how epigenetic information is propagated across cell divisions has been developed by researchers at the MRC London Institute of Medical Sciences (LMS) and the University of Copenhagen. Using this new technique, iDEMS (isolation of DNA by EdU-labelling for Mass Spectrometry), the team were able to show that only a relatively small fraction of DNA methylation is re-established immediately following DNA replication and it takes up to 12 hours to fully reinstate the original epigenetic state. Understanding this dynamic will help us to elucidate the loss of epigenetic memory observed in many human pathologies, including cancer.

All cells in the human body share the same DNA sequence, which is formed of genes that code for different proteins. Proteins carry out specific functions in cells, however, not every protein that is coded for in the DNA sequence is required for cells to carry out their function. To resolve this, chemical modifications are added to the DNA sequence to ensure only the relevant genes are active and thus only the required proteins are produced in each individual cell type. These DNA modifications are known as “epigenetic modifications”.
Epigenetic modifications can switch genes ‘on’ and ‘off’ – either activating the genes so that the proteins they code for continue to be produced in the cell, or silencing them to stop protein production. These DNA modifications include DNA methylation (5-methylcytosine) and hydroxymethylation (5-hydroxymethylcytosine), which can influence gene expression by adding methyl (CH3) or hydroxymethyl (CH2-OH) molecules to Cytosine, one of the building blocks of DNA.
Epigenetic modifications play a crucial role in normal growth and development, as well as regulating and “memorising” a response to our environment. A loss or mis-regulation of these modifications has been shown to be prevalent in genetic disorders and in many human pathologies including cancer. Due to its widespread importance in the regulation of cellular activity, epigenetics remains a fast growing area of research, and understanding how epigenetic modifications are controlled is pivotal to unlocking our understanding of disease initiation, progression and prevention.
One of the key features of DNA methylation is that it occurs on both complementary DNA strands (i.e. both strands of a DNA double helix) in a symmetric way. During DNA replication, a process that duplicates DNA in preparation for cell division, the pattern of methylation needs to be copied from the old, parental, to the newly synthesised DNA strands in order to maintain the epigenetic information across cell divisions.
Historically, it has been thought that DNA methylation is copied very quickly following the synthesis of the new DNA strands. Previous studies have attempted to characterise the exact kinetics of this process, however, contradictory results have meant that this remains a heavily debated area of science. To address this question the LMS Reprogramming and Chromatin group led by Professor Petra Hajkova and Professor Anja Groth’s group at the University of Copenhagen developed iDEMS, a highly sensitive, quantitative mass spectrometry-based approach to measure DNA modifications in the context of DNA replication.
‘iDEMS is a new tool to study the restoration of DNA modifications after DNA replication, revealing the complexity of maintaining our epigenetic memory.’ – Dr Cristina Requena, co-lead author of the publication
Using iDEMS in mouse embryonic stem cells, the team discovered that most DNA methylation marks were restored on the daughter strand within 4 hours of DNA replication. However, it took up to 12 hours post replication to fully restore the original methylation status. This suggests that the full restoration of the DNA methylation pattern takes longer than one cell cycle, raising questions about potential gradual loss of DNA methylation in fast dividing cells such as cancer cells.
This novel method also allowed to quantitatively measure the post-replication restoration of DNA hydroxymethylation levels – a question that was not possible to address in previous studies. Intriguingly, the team observed that DNA hydroxymethylation remained asymmetrically distributed between the DNA strands after replication, with the parental strand constantly showing considerably higher levels of the mark. This finding was truly unexpected and demonstrates a molecular mechanism that can be used by the cell to distinguish the original inherited DNA strands.
Summarising, iDEMS provides a new quantitative tool to address outstanding fundamental questions in the DNA replication and epigenetics fields related to how epigenetic information is propagated across cell divisions. This is of particular interest in the context of pathologies or aging, processes that have been associated with the erosion of the epigenetic memory.
This research was published in Nature Cell Biology and was led by Professor Petra Hajkova’s group at the LMS and Professor Anja Groth’s group at the University of Copenhagen. This research was funded by the Medical Research Council, European Research Council, Lundbeck Foundation, the Independent Research Fund Denmark, and the Novo Nordisk Foundation.
This news article was written by Ester Seseri.
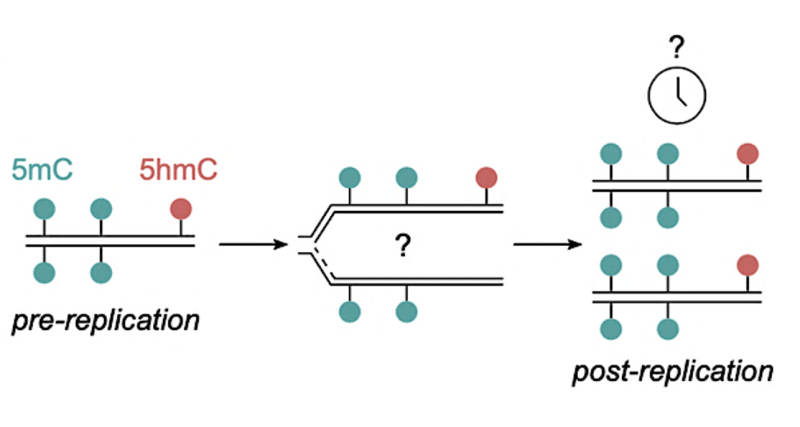 |
Post-replicative methylation maintenance. The time it takes for DNA methylation (5mC) and hydroxymethylation (5hmC) marks to be restored on the newly-formed DNA strands post-replication has been subject of intense debate. |