Researchers have produced a database and new analysis platform which could help transform drug development for difficult-to-drug diseases.
By LMS Staff Member
July 18, 2023
Time to read: 4 minutes

Researchers have produced a database and new analysis platform which could help transform drug development for difficult-to-drug diseases.
Professor Edward Tate, Professor Jesus Gil, and Matt White from the Department of Chemistry at Imperial and the MRC LMS developed a better understanding of how covalent inhibitor drugs bind to target proteins, making drugs previously thought ‘too dangerous to use’ far safer.
Covalent inhibitor drugs bind tightly to specific sites on proteins in the body which act as drug targets. Unlike other drugs, they can possess an irreversible mechanism which provides several advantages. Covalent drugs have made a major impact on human health, including the first approved drug for severe alopecia and many difficult-to-drug cancers.
Covalent drugs have made a major impact on human health, including the first approved drug for severe alopecia and many difficult-to-drug cancers.
Until the 2010s, covalent drugs were widely considered unsafe to use. Recent years have seen a spike in interest in developing covalent drugs, after finding covalent drugs could be both safe and extremely effective. This has led to a shift in the pharmaceutical industry, as more companies are investing in covalent drug research to develop new treatments.
Professor Edward Tate, who led the research, explained:
“Covalent drug discovery in combination with machine learning are rapidly emerging as critical tools to tackle a new generation of highly challenging drug targets, with many biotech and pharma companies investing heavily in this space in recent years.”
The research was primarily done at Imperial Molecular Sciences Research Hub on Imperial’s White City Campus
Drug development programs often screen potential drugs to find those with the highest specificity and effectiveness. One popular way to do this is to examine how drugs best fit into the drug target.
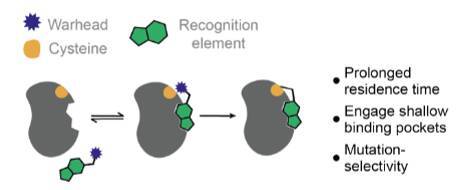
This new research, published in Cell Press, examines the cysteine groups on the surface of proteins, which can act as effective sites where drugs can bind. Artificial intelligence, proteomics and millions of data points from previous studies were used to investigate drug-protein target interactions involving cysteine groups.
Researchers successfully mapped millions of drug-drug target interactions, allowing them to visualise 95% of the cysteine side chains which the drugs could bind to. They then compiled this into an open resource that will aid drug developers in optimising covalent inhibitors/drugs they develop, making these drugs safer at even the earliest stages of development.
“Researchers successfully mapped millions of drug-drug target interactions, allowing them to visualise 95% of the cysteine side chains which the drugs could bind to.”
The team explained that their extensive database will help identify biases in sampled cysteines and provide a framework for benchmarking coverage of druggable cysteines in future cysteine profiling approaches.
We live in an exciting time for drug development with new technology, such as artificial intelligence, advancing faster than ever. This new publication marks another significant step toward maximising the use of powerful covalent drugs in safely treating even the most difficult diseases.
“Covalent inhibitors have had significant success in the clinic in the past 10 years and represent a very promising therapeutic strategy for difficult-to-drug targets. Our findings help to better understand where covalent inhibitors bind and how selective they are, as well as providing future directions for technology developments.”
Matt White, the first author of the publication
“In this paper, we have shown how meta-analysis of millions of protein-ligand interactions and hundreds of thousands of potential drug target sites can be combined with cutting edge in silico protein structure prediction to deliver new insights into covalent ligand optimisation, and a comprehensive open access database which will help to accelerate discovery against currently intractable drug targets.”
Professor Edward Tate, who led the research
Click here to access the database
This article was funded by Pfizer Ltd, Kura Oncology, Daiichi Sankyo, Oxstem, Exscientia, Myricx Pharma, AstraZeneca, Vertex Pharmaceuticals, GSK (GlaxoSmith Kline) and ADC Technologies. EWT owns equity in Myricx Pharma, Exactmer and Samsara Therapeutics, Imperial College London and the Francis Crick Institute.
Graphical Abstract of the publication