Scientists from the MRC Laboratory of Medical Sciences (LMS) have become among the first to study the initiation of human DNA replication. By pioneering new methods to isolate human proteins involved in DNA replication, the team has identified several key differences between the process in humans compared to yeast and a unique binding interface between two key proteins that can be mutated in cancer.
By Tom Wells
January 22, 2025
Time to read: 4 minutes
DNA replication is a fundamental process that ensures each time a cell divides, the two daughter cells inherit the same DNA as the parent cell.
DNA licensing is an essential first step in DNA replication that ensures a section of DNA is only copied once. If extra copies are made, this DNA can insert itself into the genome, causing mutations and disease.
During DNA licensing, multiple proteins assemble and function to load an enzyme called DNA helicase onto the DNA. This acts as a kind of molecular ‘flag’ that the DNA is ready for replication. The helicase then acts like a motor, moving along the DNA and unwinding it into two strands so that the molecular machinery required for DNA replication can access the DNA and begin copying.
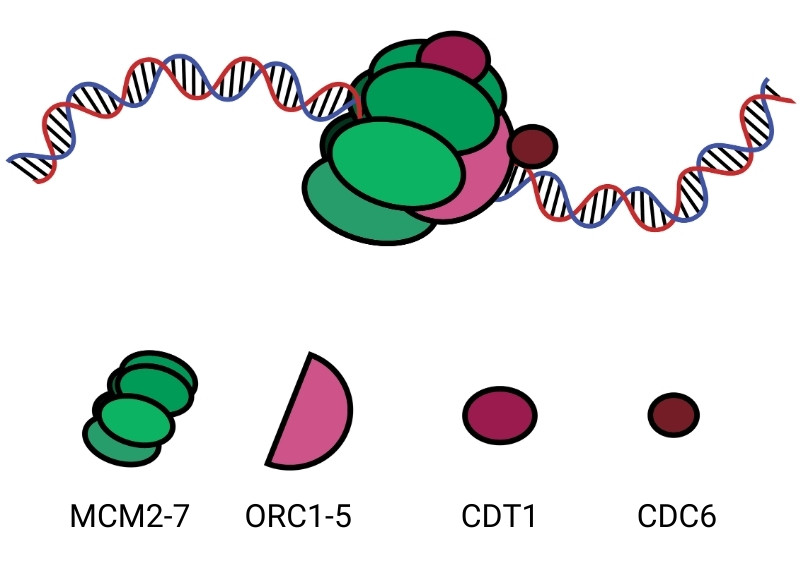
The complex which loads the helicase onto the DNA is made up of multiple proteins. These include:
Studying DNA licensing in humans was not possible until recently, due to the difficulty in isolating the proteins from human cells. Therefore, most research into DNA licensing has been done in yeast.
Using a newly developed method from the DNA replication group at the LMS, headed by Professor Christian Speck, postdoctoral researcher Jennifer Wells and PhD student Lucy Edwardes have become amongst the first researchers to be able to characterise DNA licensing in human DNA replication. The work was published earlier this month in Nature Communications, around the same time as related studies were published in Nature by Franziska Bleichert’s group at Yale University and Alessandro Costa and John Diffley’s groups at the Francis Crick Institute, which independently found methods to isolate human proteins.
Studying these proteins, the team made the surprising discovery that while the complexes of both species contained the same proteins, the order the proteins are assembled in differs between yeast and humans. This work provides a strong basis for better understanding how DNA replication is regulated.
“We wanted to discover how the first step of human DNA replication works so that we could understand disease-associated mutations,” says Christian. “We’re among some of the first researchers to successfully reconstitute the process in the test tube with purified proteins, which allowed us to understand how the process works.”
In addition, the team used an advanced microscopy technique called cryo-EM to generate a high-resolution structure of the complex. Using this structure, they were able to identify a unique interface between two of these proteins that can be mutated in cancer. The structure revealed key structural changes in ORC2 upon DNA binding, which allowed binding of Cdc6 and Mcm3. The team were intrigued by this binding and looked at the parts of the proteins that made up this interface between the helicase subunit MCM3 and CDC6. Mutations in these areas were commonly found in cancer patients and were associated with poor DNA licensing. By continuing to study this new system, researchers could learn more about mutations that cause disease which could help lead to near therapies in the future.
“Many members of the DNA Replication Group played a crucial role in this study. It’s been a privilege to work as a collaborative team on such an exciting project,” concludes Lucy. “We’re looking forward to understanding how DNA replication is misregulated, particularly in cancer and ageing.”
This work was funded by Cancer Research UK and the Wellcome Trust.
Wells, J.N., Edwardes, L.V., Leber, V. et al. Reconstitution of human DNA licensing and the structural and functional analysis of key intermediates. Nat Commun 16, 478 (2025). https://doi.org/10.1038/s41467-024-55772-z
Published 8 January 2025