By Helen Figueira
October 17, 2012
Time to read: 4 minutes
 Imaging Single Molecules
Imaging Single Molecules
Biological processes are rarely simple. The molecular pathwaysfundamental to life are intricate, complex, and might occur a thousandtimes over every moment. One way to peer into the delicate steps from Ato B might be to isolate individual molecules, and watch how theychange over time. That’s what David Rueda, the latest addition to thegrowing Integrative Biology Section at the CSC, is trying to do.
“If you do an experiment in a test tube, you’ll have an Avogadro number of molecules, each doing slightly different things,”explains Rueda, “so on average, you may see nothing happening. You lose mechanistic information. But if you take one molecule at a time, youcan follow the pathway precisely, in real time.” In the newly formed Single Molecule Imaging Group, Rueda plans to apply this philosophy to abroad range of biological quandaries.
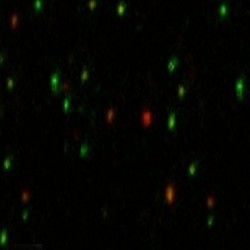
For example, the DNA-modifying enzyme APOBEC3G plays a key role in defence against retroviruses by introducing purposeful mutations[specifically, deaminating ‘C’s to ‘U’s]. “The question,” says Rueda,“is how does the enzyme locate its specific targets on the DNA? We’d like to see that in real time. So we immobilise the DNA on a microscope slide and label it with a green fluorophore. We then purify the protein and label it with a red fluorophore.” This video shows the result:
Imaging Single Molecules
Biological processes are rarely simple. The molecular pathwaysfundamental to life are intricate, complex, and might occur a thousandtimes over every moment. One way to peer into the delicate steps from Ato B might be to isolate individual molecules, and watch how theychange over time. That’s what David Rueda, the latest addition to thegrowing Integrative Biology Section at the CSC, is trying to do.
“If you do an experiment in a test tube, you’ll have an Avogadro number of molecules, each doing slightly different things,”explains Rueda, “so on average, you may see nothing happening. You lose mechanistic information. But if you take one molecule at a time, youcan follow the pathway precisely, in real time.” In the newly formed Single Molecule Imaging Group, Rueda plans to apply this philosophy to abroad range of biological quandaries.
For example, the DNA-modifying enzyme APOBEC3G plays a key role in defence against retroviruses by introducing purposeful mutations[specifically, deaminating ‘C’s to ‘U’s]. “The question,” says Rueda,“is how does the enzyme locate its specific targets on the DNA? We’d like to see that in real time. So we immobilise the DNA on a microscope slide and label it with a green fluorophore. We then purify the protein and label it with a red fluorophore.” This video shows the result:
Each green dot represents a single molecule of DNA. When a red lightoverlaps, the protein is binding to the DNA. “It works like a carantenna. The fluorophore on the protein is the receptor, and thefluorophore on the DNA the emitter. When the protein comes in closeproximity, energy is transferred from one to the other.” Measuring theintensity of the resulting red to green ratios reveals the proteinmobility. “We can then change parameters such as ionic conditions,sequence, etc, and analyse the data. That helps us tease out themolecular mechanism by which the enzyme scans the DNA, finds itstarget, and deaminates it.”
The same technique can be applied to almost any biological complex,provided Rueda and his team can purify the components. For example, RNAfolding presents a unique challenge, he explains: “RNA molecules canact as enzymes, but require very specific 3-dimensional structures tobe active. But RNA is based on just 4 bases, rather than 20 aminoacids, and those 4 bases find ways to contort into an almost limitlessnumber of forms.” Rueda labels the RNA at opposing ends, and as themolecule folds, the changing distance between the visible spots revealsthe folding reaction.
While applying the technology as it exists provides no shortage ofavenues for investigation, Rueda is also hoping to take the methodsfurther while at the CSC: “We’d also like to explore new frontiers. Sofar we’ve worked mostly in vitro, with purified components andsynthetic nucleic acids. Now we’d like to see these processes in thecell.”
AL