By Staff Member
December 1, 2023
Time to read: 6 minutes
An LMS collaboration between Professor of Imaging Sciences Declan P O’Regan and James Ware a Professor of Cardiovascular and Genomic Medicine has resulted in a novel genetic map of Hypertrophic Cardiomyopathy (HCM). This research could help to understand the genetic drivers of disease diversity and offer more personalised clinical profiles for a disease that affects one in 500 people. By providing a clearer understanding of HCM’s genetic basis, mapped to its variable physical characteristics, this paves the way for future research on why this disease can be so diverse in different people.
Hypertrophic cardiomyopathy (HCM) is linked to sudden death in early life and middle age. Usually caused by inherited genetic variants found in 1 in 500 people, HCM is characterised by an abnormal thickening (hypertrophy) of the heart muscle of the left ventricle, the main pumping chamber of the heart. The muscle walls become thick and stiff over time. HCM can lead to serious health conditions, that include abnormal heart rhythms and sometimes cardiac arrest.
HCM has long posed challenges to clinicians due to its diverse manifestations in patients. HCM may have several different patterns of wall thickening, as well as different genetic risk factors and environmental effects. The interaction of different genetic variants and patterns of disease has been unclear and relied on subjective interpretation of cardiac imaging.
According to Dr Lara Curran, stated: “HCM, a condition intricately linked to sudden cardiac death, exhibits a wide array of physical manifestations and genetic underpinnings. The traditional classification systems fall short in capturing this extensive variability, highlighting the necessity for a more sophisticated method. By employing machine learning, we’ve developed an innovative ‘tree-like’ structure for HCM categorization. This model portrays the disease’s expression as a spectrum, with distinct ‘branches’ signifying groups of patients with similar heart structures. Our work lays the groundwork for in-depth analysis of the relationships between genes and physical traits, and it advances the personalized assessment of cardiac risk.”
Recent advances have made it possible to use AI to combine and analyse huge data sets derived from patients’ individual genomes, in combination with other clinical data and data from MRI imaging of the heart, allowing them to decode the complexities of HCM and relate physical and genetic data. The researchers studied a cohort of 436 HCM patients, using additional genetic data from UK Biobank as a control, and data from a Singaporean study for external validation, to leverage a vast pool of clinical, genetic, and imaging data. Utilizing machine learning, the team scrutinized the three-dimensional structure of the heart’s left ventricle via cardiac magnetic resonance imaging (CMR), crafting a sophisticated tree-based model that could delineate the highly variable physical characteristics of HCM in different patients.
This novel approach, which blends AI’s pattern-recognition capabilities with advanced cardiac imaging, analysed over 400 patient scans to construct a 3D heart model which demonstrated a branching development of severity across variable forms of HCM. The resultant ‘family tree’ vividly maps the interplay between genetic and physical characteristics phenotypic, advancing our understanding of how the disease varies among individuals.
The new taxonomy, which is a new classification system, identifies four main phenotypic branches of HCM, which are distinguished by distinct patterns of heart muscle thickening. This classification is a significant leap from the traditional two-dimensional and subjective approaches, offering a more detailed understanding of the disease.
For patients, this research could lead to a more personalized approach to HCM management, potentially leading to more effective treatment strategies paving the way for personalised, precision medicine in the future, as it will allow clinicians to more accurately identify cardiomyopathy risk from 3D heart images. “Understanding the diverse ways this disease manifests are crucial for developing targeted treatments,” explained Declan, “the study also opens avenues for applying similar methodologies to other cardiovascular diseases, paving the way for broader advancements in personalized medicine”.
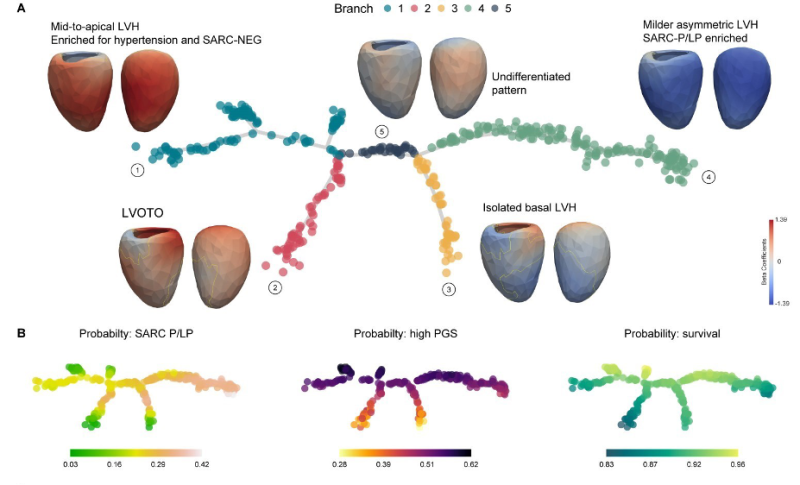
This ‘family tree’ graphic offers a striking visualization of HCM’s diversity, distilling the complex interplay of genetics and disease presentation across a cohort of roughly 500 patients with various presentations of HCM and different genetic profiles. It serves as a roadmap to the intricacies of HCM, charting the varia bleed outcomes with the patients’ genetic data and predispositions. By placing individuals within this detailed framework, the map allows clinicians to pinpoint a patient’s unique position in the landscape of the disease.
Branch 1: Mid-to-top left ventricular hypertrophy (LVH), common in patients with high blood pressure and without mutations in sarcomeric (muscle) protein. (SARC-NEG).
Branch 2: Features patients with LVH associated with obstruction of the outflow tract from the left ventricle (LVOTO).
Branch 3: Isolated thickening at the base of the heart.
Branch 4: Milder asymmetric LVH with a higher presence of sarcomeric mutations (SARC-P/LP
Branch 5: Represents an undifferentiated pattern, indicating a baseline from which other phenotypes diverge.
This study not only sheds new light on this complex heart condition but also exemplifies the power of interdisciplinary collaboration and advanced AI technology in medical research. Such innovative approaches are set to play a crucial role in shaping the future of medicine.
This finding presents the data-driven taxonomy of HCM, identifying patient groups with similar heart morphology. This taxonomy a offers continuum of disease severity, genetic risk, and outcomes, offering valuable insights into the disease’s diversity causes and consequences.
The team plans to apply these approaches to understand human diversity in a broader range of cardiovascular diseases.
In a move to encourage widespread access and further research, the authors have applied a creative commons attribution license to their research to understand human diversity in a wider range of cardiovascular diseases.
The study, supported by the Medical Research Council, National Institute for Health Research, and the British Heart Foundation, Special acknowledgments were made to Dr. Tim Dawes and Dr. Carlo Biffi from Imperial College London for their initial development of the 3D cardiac modelling, and Dr. Jinming Duan from the University of Birmingham for his work on the segmentation pipeline.
Read the paper here.