In a groundbreaking study, researchers led by PhD candidate Imanol Duran at the Medical Research Council Laboratory of Medical Science, in collaboration with Imperial College London and DKFZ in Germany, have developed machine learning algorithms to accurately identify senescent cells. This advancement promises to revolutionise treatments for cancer and aging by facilitating the development of therapies targeting senescent cells.
By Staff Member
February 14, 2024
Time to read: 6 minutes
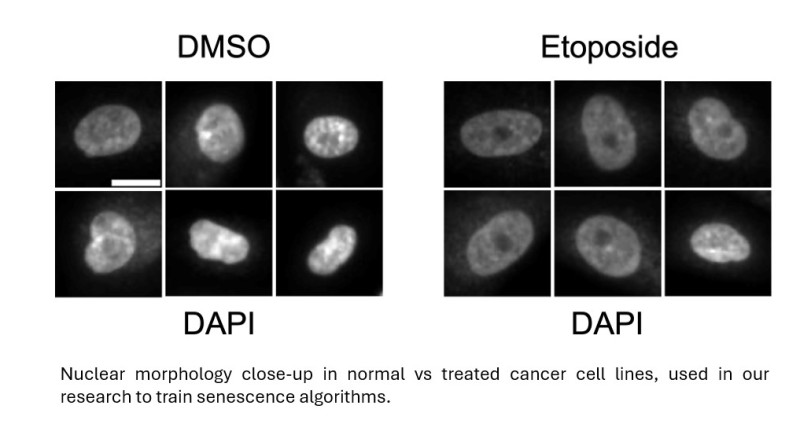 It is estimated that for a cell to become cancerous it needs to accumulate around six mutations, with those usually occurring during cell division. As cells get older, the risk of mutations occurring during this replication process also increases and for many years scientists wondered how we managed to live for many decades without accumulating genetic damage causing more cancers across a typical lifetime. Part of the answer emerged in the 1960s when Leonard Hayflick discovered that cells growing in labs could only undergo around 50 replications before they would simply stop dividing.
It is estimated that for a cell to become cancerous it needs to accumulate around six mutations, with those usually occurring during cell division. As cells get older, the risk of mutations occurring during this replication process also increases and for many years scientists wondered how we managed to live for many decades without accumulating genetic damage causing more cancers across a typical lifetime. Part of the answer emerged in the 1960s when Leonard Hayflick discovered that cells growing in labs could only undergo around 50 replications before they would simply stop dividing.
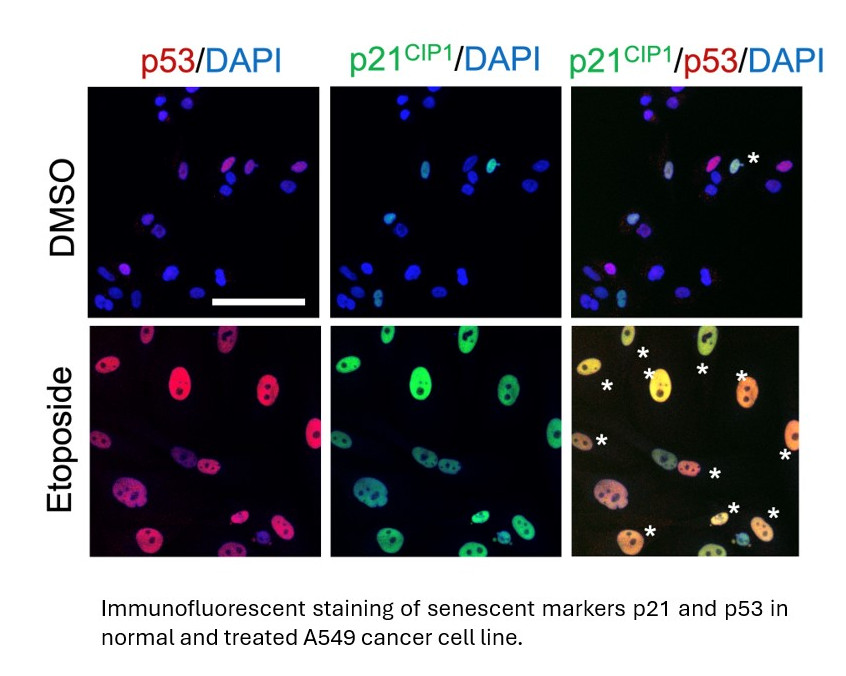 This phenomenon – that cells could only divide a certain number of times before stopping, became known as the Hayflick Limit and showed that individual cells, like their more complex human form, were mortal. It was then proposed that preventing cell replication is an integral aspect of ageing – a trade-off for averting cancer in the early decades of life. The process of cells reaching the point at which they stopped dividing was termed senescence (“senex” from the Latin “to grow old”) and occurs naturally after a cell’s Hayflick limit has been reached. It is also induced by a variety of cellular insults and stressors. Importantly, most cancer therapies also induce senescence, and their efficacy is in part related to senescence limiting tumour division.
This phenomenon – that cells could only divide a certain number of times before stopping, became known as the Hayflick Limit and showed that individual cells, like their more complex human form, were mortal. It was then proposed that preventing cell replication is an integral aspect of ageing – a trade-off for averting cancer in the early decades of life. The process of cells reaching the point at which they stopped dividing was termed senescence (“senex” from the Latin “to grow old”) and occurs naturally after a cell’s Hayflick limit has been reached. It is also induced by a variety of cellular insults and stressors. Importantly, most cancer therapies also induce senescence, and their efficacy is in part related to senescence limiting tumour division.
Therefore, induction of senescence is initially a protective response that prevents older, damaged or cancerous cells from replicating and perpetuating damage in the tissues. Normally, these senescent cells will be eliminated by the immune system. But when that does not happen and senescent cells persist in the body, far from being passive or sleeping, they behave like “zombie” cells, remaining active, even if not dividing, trading cellular insults with its immediate environment and surrounding cells, and crucially, acting as a focal point for the chronic inflammation that is a hallmark of the ageing process itself and can drive cancer progression.
Although vast strides are being made in senescence research with incredibly promising results, progress has been hampered by a lack of reliable ways to determine senescence. Virtually all cell types can be made to enter a senescent state however what senescence looks like at a cellular level, and how it can be reliably identified across thousands of cell types has been far from easy to determine. Senescent cells have many characteristics: they stop proliferating, start secreting molecules to communicate with their surrounding cells (known as the senescence-associated secretory phenotype, SASP), reprogram their metabolism and undergo changes in their cellular and nuclear morphology. However, the heterogeneity of senescent cells, combined with a lack of universal biomarkers and technical issues make the detection of senescence a bottleneck in the field. This has been recently highlighted in a commentary by the Senescence group at LMS published in Nature Cell Biology (Gil, Nat Cell Biol 2023).
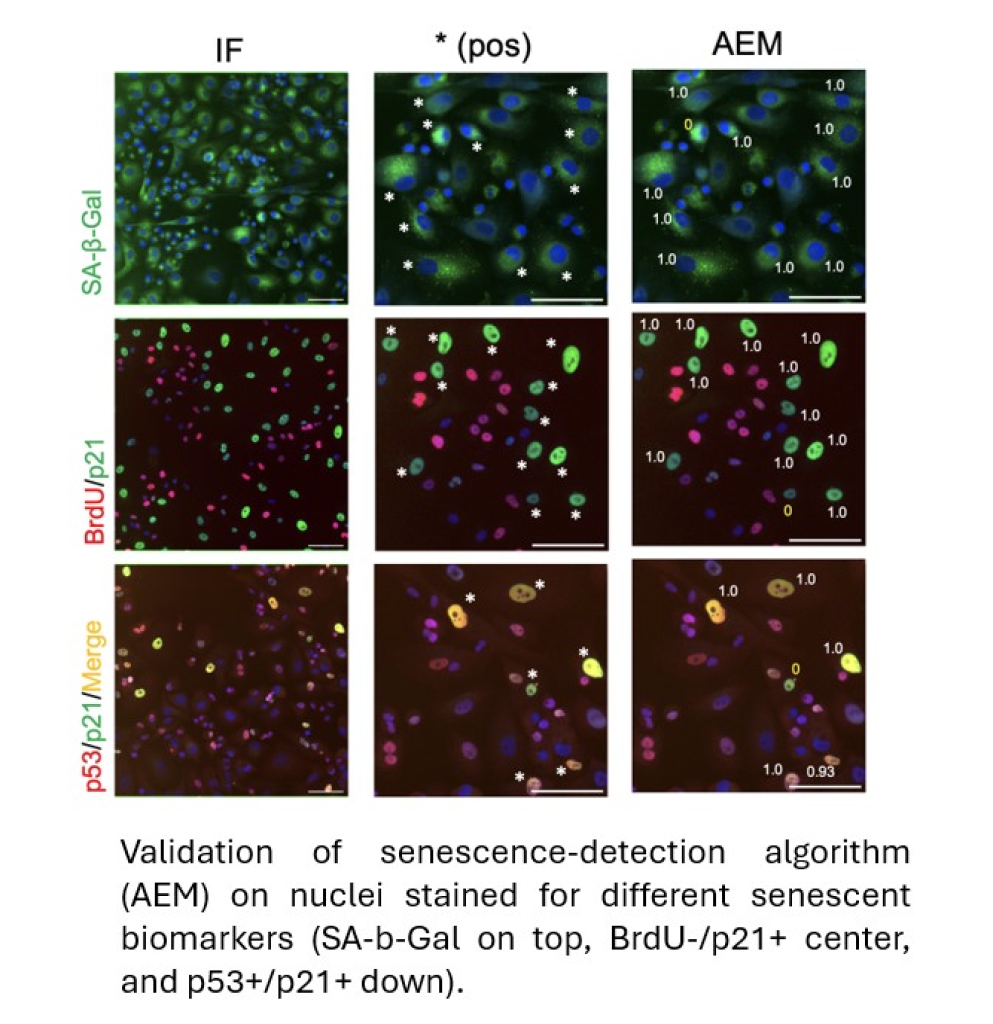
Recent advances in machine learning and AI have allowed researchers at the LMS to analyse hallmarks of senescence which can now be used to confirm the presence of senescent cells of any origin across various cell types. Not only this but they have also shown that these algorithms can be reliably adapted to work in lab-grown cells for research but also in tissue samples obtained from mouse livers and patients with fatty liver disease.
The breakthrough by PhD candidate Imanol Duran working at the Senescence Group at the MRC LMS occurred when they began looking at changes in the nuclear morphology and chromatin (DNA packaging) architecture of senescent cells in tissue culture. Although these features alone were not enough to classify or identify cells as being senescent, a detailed examination and measurement of nuclear geometry including surface area, elongation, compactness, and form factor provided a library of data which could then be subjected to machine learning. All the features were distinct in senescent cells from normal cells and Duran was able to devise a family of algorithms that detect a wide range of senescent cells, from cancer cells to primary fibroblasts, with high accuracy, using data obtained from open-source image analysis software.
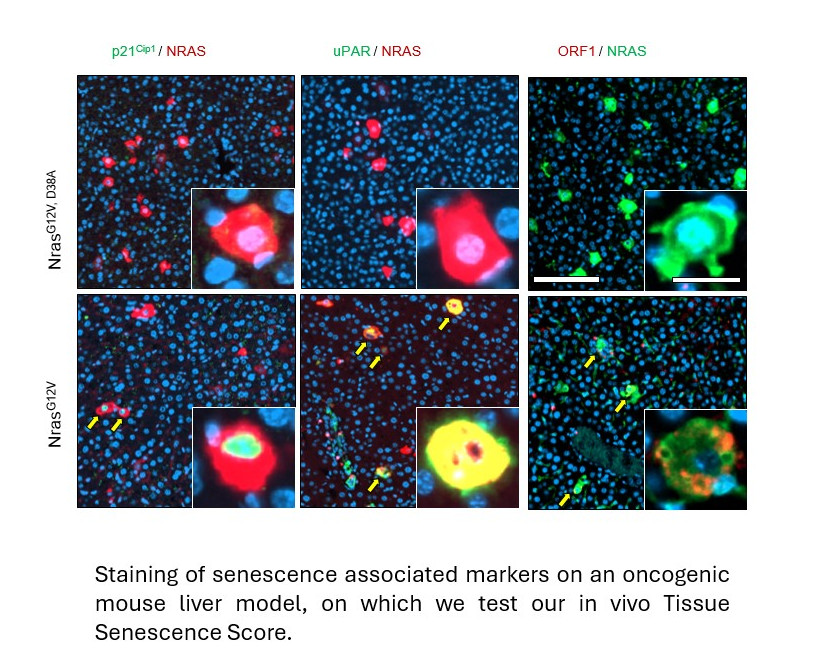
After establishing that the algorithms were robust and accurate across a range of cell types, the team moved their work to animal models and patient samples to see whether this was a tool that could be useful in clinical applications (collaborating with researchers at the Department of Metabolism at Imperial College). Excitingly, they discovered that they could use the methods to predict the progression to senescence in mouse models of cancer development and in ageing mice and in patient samples.
Senescent cells have recently been identified as potential therapeutic targets in both anti-cancer treatments, and in slowing down the ageing process – both big focuses of international research efforts. In the former, inducing potentially cancerous cells to enter senescence, could stop tumour growth in its tracks, whereas in the latter, identifying and eliminating the inflammatory senescent cells that have accumulated as part of the ageing process could slow it down. Preliminary experiments in animal models have shown that the lifespans and healthspan of animals are increased when subjects are treated with so-called ‘senolytic’ drugs that attack and kill senescent cells.
 The researchers at LMS discovered that they could use their classifiers of senescence to identify drugs that either kill senescent cells (senolytics) or selectively induce senescence in cancer cells in vitro. In addition, the tool enabled them to actively and quickly screen a catalogue of over 600 drugs for senolytic activity, identifying several new potential therapeutic drugs for future use.
The researchers at LMS discovered that they could use their classifiers of senescence to identify drugs that either kill senescent cells (senolytics) or selectively induce senescence in cancer cells in vitro. In addition, the tool enabled them to actively and quickly screen a catalogue of over 600 drugs for senolytic activity, identifying several new potential therapeutic drugs for future use.
 Given that the accurate identification of senescent cells has been a bottleneck to progress in this fast-moving and high-impact area of research, opening the door for clinical use of this tool to target senescence in both cancer, and ageing takes us a big leap forward towards a new era of senolytic therapies.
Given that the accurate identification of senescent cells has been a bottleneck to progress in this fast-moving and high-impact area of research, opening the door for clinical use of this tool to target senescence in both cancer, and ageing takes us a big leap forward towards a new era of senolytic therapies.
The study was published in Nature Communications on February 3rd, 2024, as a collaborative effort of research groups at LMS, Imperial College London, and DKFZ in Germany. MRC core funding and CRUK were the main funders of this study, a full list of funders is included in the paper.
Duran et al. Detection of senescence using machine learning algorithms based on nuclear features. Nat Commun. (2024) 15:1041. Doi: 10.1038/s41467-024-45421-w.
Gil J. The challenge of identifying senescent cells. Nat Cell Biol. (2023) 25, 1554-1556.