By Helen Figueira
May 17, 2012
Time to read: 3 minutes
 Unpicking the molecular mechanisms of chromatin biochemistry
Unpicking the molecular mechanisms of chromatin biochemistry
An inspiring lecturer speaking about gene silencing in yeast sparked Till Bartke’s fascination with gene regulation. Having completed his PhD at the Max Planck Institute of Biochemistry, he let his curiosity about chromatin’s role in gene regulation lead him to Cambridge for his postdoctoral research. He has now brought his research to the CSC, and formed the Chromatin Biochemistry group.
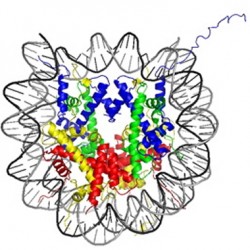
“The total length of DNA in every cell is about 2 metres, and it is all crammed into a tiny nucleus,” explains Bartke. “In order to do this, it is folded up at several levels. At the basic level, it’s wound around a protein complex which consists of four histone proteins. This particle made up of histones and DNA is called a nucleosome and is the basic unit of chromatin.” This structural arrangement of DNA influences how genes are expressed. Histone proteins have short amino acid ‘tails’, which protrude out of the nucleosome particle. Enzymes in the nucleus can add or remove various modifications to these tails, which affect how the corresponding DNA is read. “These modifications can be recognised by other proteins. The question I had was how do proteins extract the information that is stored in the patterns of modifications on the nucleosome?” says Bartke.
Working in Tony Kouzarides’ Lab in Cambridge, Bartke began investigations into how to artificially construct a nucleosome, to analyse the mechanisms involved in this form of gene regulation. “It deals with synthetic biological parts. The idea was to generate modified histone proteins and fold them into complete nucleosomes. We would produce four histone proteins separately, then chemically unfold them, mix them, and put them back together.” The technique uses artificial DNA sequences with a strong propensity for wrapping around the histones. Periodic small kinks in the DNA strand help it naturally curve around the proteins, and cement interactions between DNA and histones that lead to the formation of the stable nucleosome.
Using mass spectrometry, Bartke could then analyse which proteins bind to three specific histone modifications, combined with variations in DNA methylation (a gene regulatory process that adds a methyl group directly to the DNA). “Here at the CSC, I plan to take this system and develop it further. The goal is to understand how chromatin is regulated at the molecular scale. There are two levels to my work. First, we must identify the players: a far-reaching search for the proteins involved. The second approach is to take these proteins and analyse them in molecular detail to find out how they work.”
Bartke sees tailored cancer therapy as the ultimate aim of the research. “I’ve always been interested in cancer. It has become clear that chromatin structure and regulation can play a big role in cancer, but we simply don’t know how it works. Cancers can develop when tumour suppressor genes are downregulated, and chromatin is important in this. It’s not a mutation, so it could potentially be reverted, meaning we could rescue cells. Cancer is not a simple disease, and epigenetic therapy is still in its infancy. We are far away from applicable drugs, but this is a good place to start.”
–AL