By Sophie Arthur
July 13, 2020
Time to read: 3 minutes
Each cell in our bodies has 2 metres of DNA packed inside its nucleus. The long string of DNA is wound up and tightly packed into structures called chromosomes. However, in order for a cell to be able to carry out all the basic processes it needs, the DNA structure needs to be organised and manipulated.
One of the ways that the chromosomes are structured is through the formation of large loops of DNA. These loops are formed by Structural Maintenance of Chromosomes (SMC) proteins, but exactly how these proteins fold the DNA into these fundamental 3D structures is an unresolved question of chromosome biology.
Research published today in Nature Structural and Molecular Biology, involving the Cell Cycle group at the LMS, uses a technique called cryo-electron microscopy (cryo-EM) to investigate one of these SMC proteins, condensin, and how it manipulates the DNA. Condensin helps to form compact chromosomes during mitosis – a type of cell division that results in two daughter cells that each have the same number of chromosomes as the parent cell. The chromosome architecture formed by condensin is essential in order for the daughter cells to inherit the correct number of chromosomes.
In this study, the condensin complex was purified and its structure studied by cryo-EM. Condensin, like other SMC complexes, are ATPases. They are powered by an enzymatic reaction called hydrolysis, in which the molecule ATP (adenosine triphosphate) is broken down into ADP (adenosine diphosphate). In today’s research, the team analysed the structure of condensin with ATP present, and then absent.
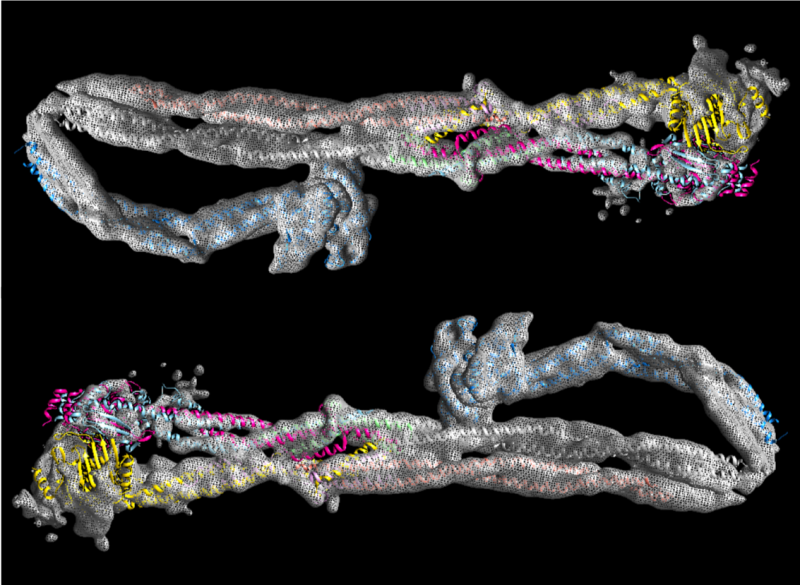
Researchers saw a change in the structure of condensin when they added ATP. This therefore suggests that ATP is likely to be critical for the activity of the condensin, and successful creation of the compact chromosomes required for successful cell division during mitosis. These structural changes could also form the basis of the action of all other SMC complexes too – which are highly conserved ATPases.
Luis Aragon, Head of the Cell Cycle group at the MRC LMS and one of the senior authors on this study said:
“This work will be a landmark in the field of nuclear architecture and chromosome biology because it shows the first condensin structure in different states. It was only possible because of the united efforts of several leading laboratories with complementary expertise.”
‘Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism’ was published on 13 July in Nature Structural and Molecular Biology. Read the full article here.