Researchers at the MRC Laboratory of Medical Sciences (LMS) have used a genomic technique called ChIP-exo to reveal for the first time how enzymes called DNA helicases get equally distributed across the whole genome in budding yeast to ensure accurate DNA replication throughout. This discovery could have implications for cancer and age-related diseases in the future.
By LMS Staff Member
August 28, 2024
Time to read: 4 minutes
DNA replication is a fundamental biological process which ensures that each time a cell divides, the two daughter cells inherit the same DNA as the parent cell. Errors in DNA replication accumulate over time, causing genetic changes that contribute to ageing and increase the risk of diseases, such as cancer.
The process begins at specific locations within the genome, called replication origins. It involves a variety of molecular components, including enzymes like DNA helicases, which unwind double-stranded DNA, and loading complexes, which help assemble the necessary molecules onto the DNA.
DNA must be accurately copied and replicated at all replication origins across the whole genome, but until now scientists haven’t understood exactly how this happens.
In research published last week in Nature Communications, researchers from the DNA Replication Group have revealed for the first time that the binding sites of origin recognition complex (ORC), a loading complex, and MCM2-7, a DNA helicase, overlap in yeast. This provides a molecular explanation for why DNA helicase assembly at origin sites can only happen once at each site, ensuring that DNA helicases are equally distributed across the genome.
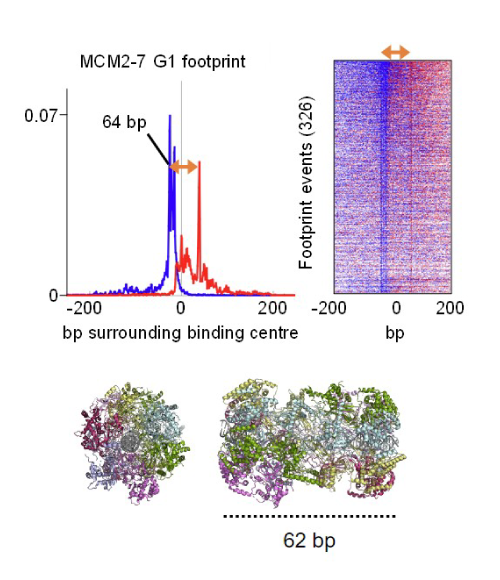
For many years, scientists supposed that multiple DNA helicases were loaded onto one origin site to initiate replication, but they lacked an experimental technique powerful enough to answer this question.
The DNA Replication Group explored this question using a technique called ChIP-exo, which allows researchers to see the complex assembly of proteins on top of strands of DNA in high resolution, to determine the exact binding sites of ORC and MCM2-7 on DNA in yeast. They found that ChIP-exo provides a similar level of detail on the DNA binding sites to that of an advanced microscopy technique called cryogenic electron microscopy (cryo-EM), which is a method used by the DNA Replication Group.
The group discovered that MCM2-7 loading leads to displacement of the helicase loader (ORC), which does not happen when a helicase loader co-factor or the helicase becomes inactive.
Further in-depth analysis in the publication shows that the primary and secondary MCM2-7 loading sites are close to each other, contain unique structural deformations and showcases that replication origins in yeast are highly compact.
This foundational research in yeast sheds light on the once mysterious mechanism that allows DNA to be accurately replicated across the whole genome.
Each component of DNA replication is essential, meaning this discovery could be targeted to develop novel treatments in the future. For example, cancer cells are highly sensitive to the inhibition of DNA replication, and there could therefore be great potential for developing novel DNA replication inhibitors against this process.
Professor Christian Speck, Head of the DNA Replication Group and senior author said: “This study exemplifies how curiosity-driven research can uncover some of the most fundamental cellular processes. By applying a cutting-edge genomics technique, we have revealed basic principles of DNA replication in yeast that could one day be exploited to engineer novel DNA replication inhibitors for cancer and age-related diseases.”
The team now plans to study the process in human cells to further enrich our understanding of DNA replication.
This work was led by Dr Max Reuter, former postdoctoral fellow in the DNA Replication Group. It was supported by the Biotechnology and Biological Sciences Research Council (BBSRC), the Wellcome Trust and the Medical Research Council (MRC).
Read the full publication: Reuter, L.M., Khadayate, S.P., Mossler, A. et al. MCM2-7 loading-dependent ORC release ensures genome-wide origin licensing. Nat Commun 15, 7306 (2024). https://doi.org/10.1038/s41467-024-51538-9