An interdisciplinary collaboration between researchers at the MRC Laboratory of Medical Sciences (LMS) and Imperial College London has revealed for the first time how a fundamental enzyme called SWR1 ‘flips’ its substrate, a protein-DNA complex called the nucleosome, to exchange protein subunits within it. Modifying nucleosomes is vital for activating many genes and signalling DNA damage for repair.
By LMS Staff Member
November 6, 2024
Time to read: 4 minutes
Research published today in Nature has revealed how the SWR1 enzyme works in yeast. This work is the result of a longstanding collaboration between Professor David Rueda and Dr Paul Girvan from the Single Molecule Imaging group at the LMS and Professor Dale Wigley FRS and Dr Adam Jalal from the Department of Infectious Disease at Imperial College London.
SWR1 is a key part of the machinery that organises DNA within cells and is important to all multicellular life, being present in both humans and yeast. Mutations in the human equivalent of SWR1, termed SRCAP, result in Floating-Harbor syndrome, a rare condition that causes short stature, speech and language delay, and characteristic facial features. Packaging DNA into cells is a complex task since over two metres of DNA needs to fit into each cell, while remaining accessible to the molecular machinery needed to activate genes. Nature has solved this problem, in part, through nucleosomes which are made up of eight subunits, called histones, that form a cylinder around which DNA wraps tightly to condense itself into a super-structure called chromatin. In humans, the chromatin is further condensed to form 23 pairs of chromosomes that carry our genetic information.
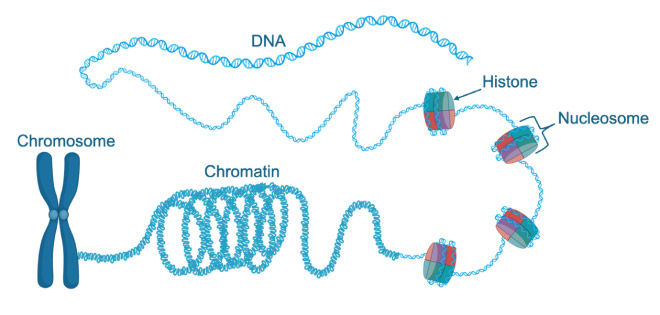
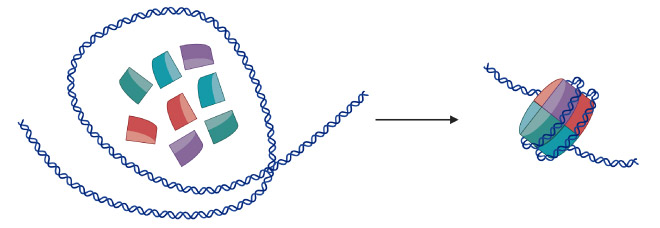
Ingeniously, nature can also use the histones in nucleosomes as signals. By modifying or varying the type of subunit in a nucleosome, this can act as a signalling system inside cells, telling the cell to activate or deactivate a gene, or highlighting a damaged section of DNA that needs to be repaired. Specifically, the SWR1 enzyme exchanges the default H2A subunit for the H2A.Z subunit, marking either the beginning of a gene or a site of DNA damage.
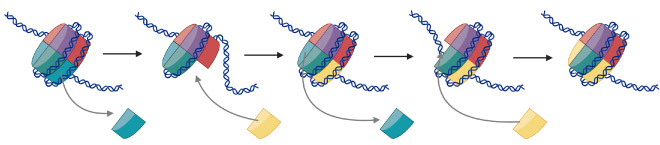
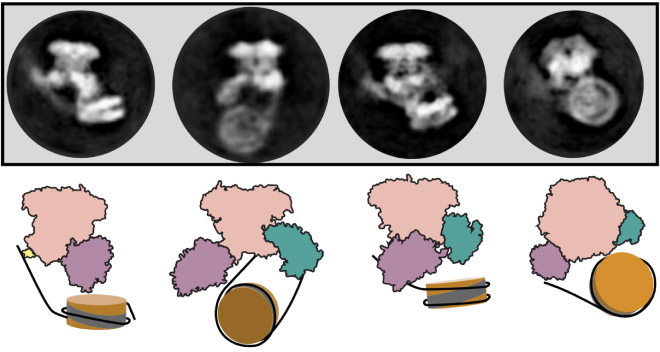
While researchers have known for many years that SWR1 is responsible for exchanging histones in nucleosomes, until now it was unclear how the SWR1 enzyme was able to modify both ends of the nucleosome successively without detaching and reattaching. By combining cryogenic electron microscopy with single-molecule fluorescence techniques, scientists have now shown for the first time that the SWR1 enzyme binds onto nucleosomes and flips them like a pancake, so that it can modify the nucleosome at both ends in a process that Paul describes as ‘molecular gymnastics’. Understanding this unusual ‘flipping’ mechanism – only elucidated through this combination of techniques, could open the door to other enzyme action discoveries.
The modifications made by SWR1 mark the nucleosome as ‘different’. Although the precise role of the modifications is not fully clear, it is known to be vital for activation of many genes and can also be a marker to signal DNA repair to damaged DNA, making the enzyme fundamental to human life.
“One of the most exciting parts of science is bringing together diverse expertise. The combination of our single-molecule fluorescent work, which showed that SWR1 was not completely unbinding from the nucleosome between exchanges, and the seeing-is-believing structural biology work that identified how SWR1 remains bound was crucial for this discovery” says Paul, “this was a real Team Science effort and developing our techniques across disciplines is shedding new light on these complex cell dynamics that help us understand the very fundamentals of life.”
This work was funded by a core grant from the MRC Laboratory of Medical Sciences, the Medical Research Council, Wellcome Trust and Cancer Research UK.
Read the full publication: Girvan, Jalal et al., Nucleosome flipping drives kinetic proofreading and processivity by SWR1, Nature (2024)