New research from the MRC Laboratory of Medical Sciences (LMS) has shed light on how regulatory regions of DNA, often mutated in diseases, switch on genes located far away from them on chromosomes. The study shows that the same DNA sequences responsible for switching these regions ‘on’ and ‘off’ also control the formation of long-distance chromosomal connections with their target genes.
By Tom Wells
January 27, 2025
Time to read: 3 minutes
DNA transcription is the vital first step needed for switching on our genes. For a gene to be switched on it must be acted upon by an enzyme called RNA polymerase. This is a molecular machine which can read DNA and create a matching strand of RNA which is then used to make proteins. RNA polymerase binds to a region just before the gene, called the promoter, before moving down the DNA to the gene.
Enhancers are regions of DNA that act as molecular switches for turning genes on or off. Understanding enhancer function is very important, since they contain the majority of genetic variants (‘mutations’) associated with common diseases, such as autoimmune, metabolic and neurological disorders. Strikingly, enhancer regions can be thousands of DNA base pairs away from the genes they control. The 3D looping of DNA brings enhancers and the promoters of their target genes into contact. To switch a gene on, the enhancer also needs to become ‘activated’ through acquiring specific chemical modifications and recruiting various proteins. However, it remains unclear whether the looping of enhancers and their activation are regulated separately or together.
New research published today in Nature Communications by the Functional Gene Control group at the LMS (Principal Investigator: Dr Mikhail Spivakov) and their collaborators suggests that a shared genetic mechanism regulates both processes.
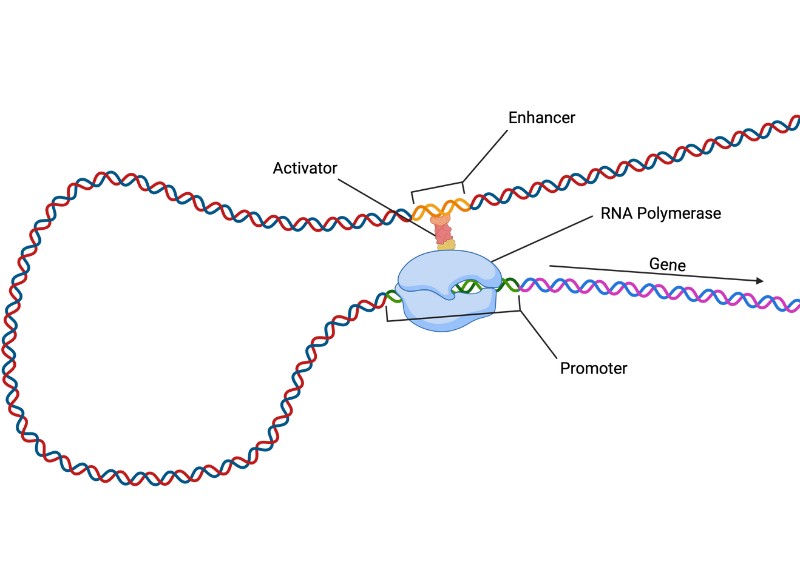
Using a combination of laboratory techniques and statistical analyses, the team studied the naturally occurring genetic variation between humans in blood cells from 34 people, focusing on DNA variants known to affect the levels of expression of specific genes, many of which were also associated with measurements of blood components, such as white blood cell count, and common diseases. They found that nearly all variants that affected the contacts between enhancers and promoters also influenced the activation of enhancers. This points to a shared genetic mechanism for the two processes during which enhancers, once activated, become ‘loopy’ and reach out to their target gene promoters. The team has validated this model using a statistical method that can infer causal relationships between phenomena and by forcibly inactivating candidate enhancers, showing that their contacts with a promoter were weakened in this setting.
“Our work demonstrates the power of using human genetic variation as ‘nature’s mutagenesis experiment’ to probe fundamental biological mechanisms,” says Mikhail, “The added advantage is that we learn about the function of variants associated with human diseases in this process.”
This work was a collaboration between researchers at the LMS, VIB Center for Molecular Neurology (University of Antwerp), NHS Blood and Transplant and MRC Biostatistics Unit (University of Cambridge) and other centres, with the Antwerp team led by Professor Valeriya Malysheva, an alumna of the Spivakov’s team.
Mikhail says: “Leveraging this huge resource of natural genetic variation for studying molecular function is both logistically and methodologically challenging. We were fortunate to have fantastic collaborators on board such as Leonardo Bottolo, Elena Vigorito and Chris Wallace from MRC Biostatistics Unit to help us with statistical genetics analysis and Mattia Frontini’s team at NHS Blood and Transplant to isolate blood cells from human donors on a large scale.”
The work was primarily funded by the Medical Research Council.
Read the full article here: Genetic coupling of enhancer activity and connectivity in gene expression control