By Sophie Arthur
August 27, 2019
Time to read: 6 minutes
When we look at our family tree, it will include hundreds of family members. Thousands if we look back far and wide enough. But each individual has a different role to play and a different story to tell, even if some of them are still a mystery to us. The same goes for our genes. We each have approximately 20,000 genes and most of them belong to a certain gene family. But in many cases, it is not clear what the job of each individual member is.
Throughout evolution, genes have become duplicated. Most of the time, these duplicated copies are lost relatively quickly because they have no useful function. But there are two ways in which a duplicated gene could become indispensable, allowing it to persist on a more permanent basis. One way, which is a very rare occurrence, is to acquire a new function that is essential to the inner workings of a cell. The other, and much more common, way is for each duplicated gene to change where it is expressed in our bodies compared to its other family members, so that each family member – also called a paralog – that has arisen from the same ancestral gene becomes specialised in one area and indispensable as a result. It is often thought that each of these paralogs, or family members, has the same function wherever it is expressed. So, in theory, you could replace one family member with another one, and the cell would still function as usual as if nothing had changed – called functional redundancy.
Research published on 26 August in Nature Immunology is challenging this idea of redundancy and asking whether these paralogs really are the same functionally.
A twist in the story of the Runx family
To test this question, researchers in the Lymphocyte Development group here at the LMS chose the Runx gene family because of the diverse expression patterns of each of its members. They were particularly interested in Runx1, which is normally expressed in immune cells called regulatory T cells, and Runx3, which is usually found in another type of immune cell called Langerhans cells.
Researchers found that when they switched which Runx family member was expressed in each cell type, the member that isn’t normally expressed in that cell type had a much stronger impact during the process of differentiation, where an immature cell becomes mature. Moreover, the foreign family member for that cell was so much more powerful at switching on genes necessary for differentiation, that it didn’t need the supporting signals that would normally be integrated in the maturation process. This suggests that the two paralogs are not as functionally redundant as first thought.
Taking a look at the ancestral tree
Having made this fascinating observation in two different cells types, the team wanted to delve a little deeper to try and work out the mechanisms behind these differences. So, the researchers used a phylogenetic approach to take a look back and find the most likely sequence of that ancestral Runx gene before the duplication events that created today’s family. In one instance, functional differences between family members were due to the sequences in a conserved domain of Runx transcription factors called the RUNT domain, where there are only 9 amino acids that are different between the RUNT domain in Runx1 and Runx3.
Asking whether these evolutionary changes in amino acids were relevant to the strength of Runx binding and subsequent activation of key differentiation genes, the team swapped certain amino acids in the sequence for their ancestral counterparts. Needless to say, the results showed that changing these amino acids did affect the strength, and as such could work out whether the ancestral sequence had stronger binding that today’s – and it did!
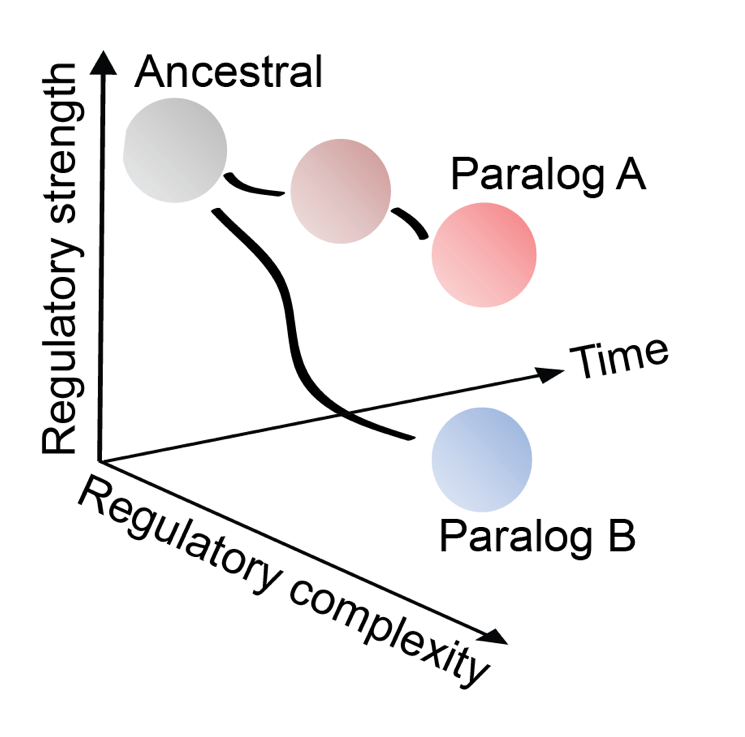
These findings highlight how stronger is not always better when it comes to regulating which genes get switched on and off in your cells. It suggests that evolution hasn’t maximised transcription factor binding strength, but in this instance modulated it to make it weaker. A possible explanation for this is that as we have evolved, we have become much more complex organisms, and by using the weaker paralog you can make differentiation more receptive to the environmental signals and allow more control over the entirety of this vital process.
First author, Ludovica Bruno, discussed her excitement about the findings:
“We were initially interested in this research because it is important for our immune system that you have that continued protection. So, we started investigating how to induce the maturation of regulatory T cells and make them stable which could help with immune diseases. But then we made this incredible observation and we wanted to explore it further to try and understand why, which gave me an opportunity to learn about many new techniques and research areas than I would have done before in order to build up this story, especially when we started looking from an evolutionary perspective.”
Matthias Merkenschlager, senior author & joint head of the Lymphocyte Development group, discussed the next steps for this work:
“Realising that we could map functional differences between the Runx members was very exciting, but more so that it was the weaker paralogs that are the ones crucial for regulating differentiation, at least in these two cell types. The next steps would be to explore whether there are more transcription factor families where this sub-maximal, but optimal, DNA binding occurs. We are also curious as to how these expression patterns have changed through evolution, so we are looking to collaborate with Peter Sarkies’ group at the LMS to look at how the regulatory elements that control transcription factor binding have changed today from the ancestral gene.”
Ludovica also wanted to highlight the support network in the team:
“I am in a position where I have no pressure to publish a paper quickly. Having that longer-term perspective has meant I have been able to invest time finding the answers to fundamental biological questions. Without that pressure, we have been able to spend the time making sense of the data and test it in more than one cell type. That would not have been possible otherwise. I have been given the respect and freedom by Matthias and in my current role I could not be happier. I also want to acknowledge the ‘silent support’ I have got from the rest of my lab peers. The ones who have helped me out which are not acknowledged on the paper for helping me learn new things, and are always willing to support not only me but the other lab members. It is a fantastic cohesive lab group to be a part of.”
‘Selective deployment of transcription factor paralogs with submaximal strength facilitates gene regulation in the immune system’ was published in Nature Immunology on 26 August. Read the article here.