By Sophie Arthur
April 30, 2020
Time to read: 6 minutes
Where did we come from? How did we become what we are? These are a few of the ‘big’ questions we ask ourselves from time to time. Although we have asked these questions for centuries, there is still lots more to explore and discover to try to answer this seemingly simple question. Researchers in the Gene Regulation and Chromatin Group have shared some new insights published in Molecular Biology and Evolution on 7 March on another of the events during our evolution that have helped us to evolve into who and what we are today.
Like all other living organisms, the vertebrates, which are the branch of life that we belong to, evolved first in water and then adapted to colonise the land. The first steps towards this occurred in the Devonian period between 400 and 360 million years ago when a subgroup of fish developed limbs, giving rise to tetrapod amphibia. This in turn led to evolution of the amniotes which first appeared in the Carboniferous period between 340 and 314 million year ago. In addition to being able to move and breath on dry land, one of the major adaptive features in amniotes was the ability of the amniote embryo to develop within a set of extra-embryonic membranes. This protects the embryo from dessication, allowing eggs to be laid and hatched on dry land, in contast to amphibia, which must return to water to reproduce.
Amniotes comprise all living mammals, birds, reptiles and their extinct relatives and ancestors. These latest findings from our colleagues have shown how a single change in the DNA of the common ancestor of all of these creatures may have contributed to the evolution of those extra-embryonic membranes that are characteristic of this group of animals. Our DNA is subject to mutations all the time, but some of those mutations can be advantageous and so become incorporated into our DNA long-term as a result of evolutionary pressure.
When our DNA is translated into proteins, it is read in threes. Each of those three letter combinations code for a specific building block of proteins called an amino acid, of which there are 20 naturally occurring ones. These amino acids are joined together to form a protein. However, when there is a change in the DNA sequence this can change the amino acid that is added at that point in the protein chain, which can have some significant consequences on the resulting protein and how it functions.
In this particular study, it was observed that an amino acid located at a specific position in an enzyme called UBE2D3 was originally an alanine, but in all living amniotes the amino acid at this exact same position is a serine. This implies that the switch from alanine to serine, which is called a substitution, occurred in the common ancestor to modern amniotes and has been conserved ever since. UBE2D3 participates in the ubiquitination of proteins, which regulates a wide range of processes in living cells
Ubiquitin is a small protein found across most cells. Ubiquitination is the term used to describe the addition of this tag, and it can affect proteins in many different ways. For example, it can mark them for degradation, or it can change their location within a cell. Other changes promoted by this versatile system include modifying signalling pathways and promoting or preventing interaction between particular proteins. The process of ubiquitination involves three main sequential steps; each with their own specific class of enzymes called E1s, E2s and E3s. UBE2D3 belongs to the E2 group. The switch from an alanine to a serine in the UBE2D3 protein created a target site for addition of a phosphate group (phosphorylation) by an enzyme called Aurora B. Phosphorylation of the serine by Aurora B introduces a negative charge, which causes UBE2D3 to unfold, making it unstable.
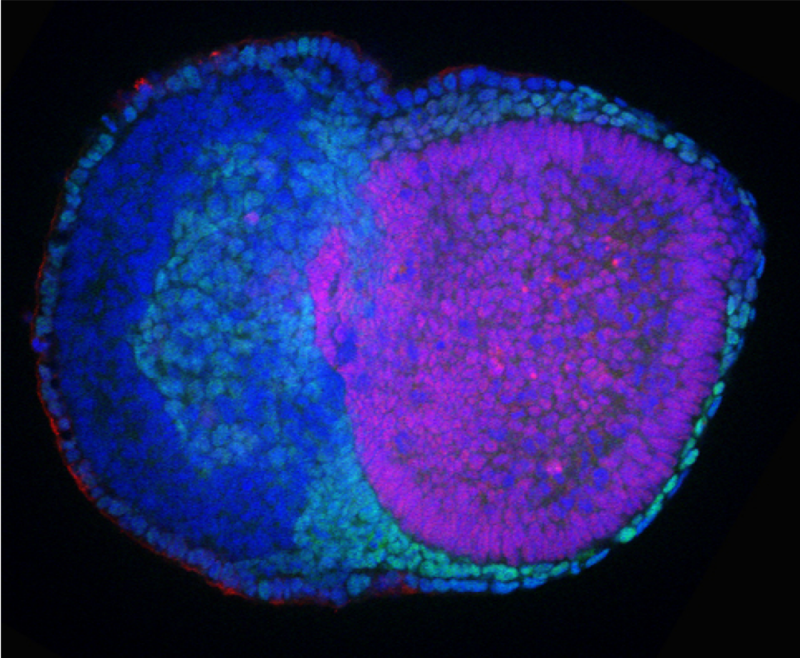
Having identified this effect on the stability of the protein, the researchers mutated the amino acid serine back to the alanine that is found in amphibia and fish. They did this using mouse embryonic stem cells and mouse embryos. What they found was that switching this amino acid back saw an increase in the E2 enzyme UBE2D3. Crucially, they found that the embryos where the amino acid had been switched to alanine didn’t survive past the stage just after implantation into the wall of the womb.
Delving a little deeper into the reason for this using mouse embryonic stem cells, the researchers found that the cells of the embryo were not able to differentiate into primitive endoderm – a layer that is crucial for formation of the yolk sac, which is important for nutrition in the growing embryo. Without the primitive endoderm, embryos cannot form visceral endoderm, which in turn guides formation of a structure called the primitive streak. This marks the start of a process in development called gastrulation which is the first stage in the formation of the tissues that make up the embryo. Without these early structures, development cannot continue. These structures didn’t form in these mouse models because the increased level of the enzyme UBE2D3, which is part of the E2 group, led it to interact more with an enzyme in the E3 group called CBL. One of the things that CBL does is to add ubiquitin to growth factor receptors, causing them to be degraded more rapidly. The downstream consequence of this was that the signals that are needed to make primitive endoderm were much lower than they should have been.
Evolution of a complex cell type such as amniote primitive endoderm will have involved many different genetic changes. The results of this study suggest that the change to the regulation of ubiquitination contributed to this evolution by altering the response of cells to signals that affect differentiation and survival of primitive endoderm cells.
Niall Dillon, Head of the Gene Regulation and Chromatin group and senior author of this study, discussed the likely next steps for research into the regulation of UBE2D3:
“It will be important to get more detailed information about what phosphorylation of the serine in UBE2D3 is doing in the early embryo. Aurora B, the enzyme that adds the phosphate group, is involved in regulating cell division and it is known that cells in mouse embryos divide very rapidly in the stage before gastrulation. We would like to know whether regulation of UBE2D3 by Aurora B is involved in speeding up cell division at this stage. Another important next step is to introduce the serine to alanine mutation in specific adult tissues and work out what else it is doing to get a broader picture of the effects of UBE2D3 regulation and how it might be implicated in diseases. This is important as we already know that the E2 enzyme has a role in leukaemia and affects the immune system”.
‘Evolution of an amniote-specific mechanism for modulating ubiquitin signalling via phosphoregulation of the E2 enzyme UBE2D3’ was published on 7 March in Molecular Biology and Evolution. Read the article here.