We explore the mechanisms underlying heart disease by applying machine learning and data science techniques to capture complex traits in the heart, across diverse populations, for discovering genetic and environmental determinants of health. Our work aims to understand the mechanisms driving diversity in heart disease.
“How can we predict cardiovascular health?”
Our work focuses on developing new techniques to learn about the heart, how it functions, and how we can better predict the effects of heart disease.
One approach to study how the heart functions is to analyse its movement through a range of images taken over time. Our work uses detailed medical imaging of heart in patients and volunteers who had had cardiac MRIs.
We develop AI algorithms to analyse the structure and function of the heart – creating a 3D digital model. We then combine clinical and genetic data with information on the motion of the heart. The builds a “digital twin” of each heart that helps us to understand how disease develops. We are applying this to up to 100,000 participants in UK Biobank as well as other large datasets of patients with cardiomyopathy.
Our aim is to pinpoint genes that influence how the develops and functions, identify risk factors for premature aging, discover potential new treatments for common heart conditions, and make personalised predictions of how health changes over time.
Part of our work is computational – utilising and developing algorithms and manipulating existing datasets.
We perform genome-wide association studies to identify potential genes that are linked to different functionalities of the heart, as well as carry out stimulations and record observational data from clinical trials.
We hope that a better understanding of genetic and environmental risk factors for heart disease will accelerate the discovery of new treatments.
“We explore the mechanisms underlying heart function using machine learning to analyse cardiac motion for predicting patient outcomes, discovering potential therapeutic targets and identifying genetic risk factors.”
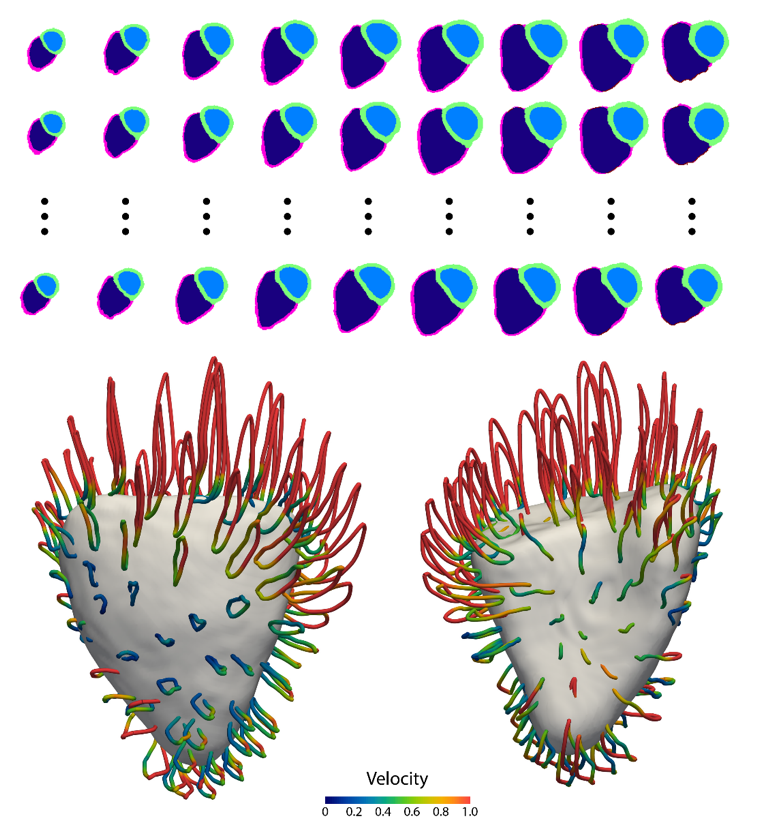
Image segmentation and motion analysis: Techniques for vision-based motion analysis aim to understand the behaviour of moving objects in image sequences. In this domain, deep-learning architectures have achieved a wide range of competencies for object tracking, action recognition and semantic segmentation. We are developing novel deep learning techniques for segmentation, motion analysis and registration of diverse cardiac MRI datasets that are efficiently scalable to large populations. This work provides a framework for quantitatively phenotyping complex cardiac traits including function, geometry and tissue characteristics for classification and prediction tasks.
Automated adverse event prediction in heart disease: Current approaches for survival prediction rely on hand-crafted features that lack precision and are insensitive to the complex pathophysiology of heart failure. We are evaluating the use of deep learning algorithms to predict time-to-events from complex input data that include motion phenotypes, genetics and clinical variables. We are also exploring how to integrate irregularly sampled data and model competing risks in heart disease as a step towards the development of artificial reasoning systems for automated analysis and interpretation of clinical investigations to optimise management decisions.
Genotype-phenotype modelling: Substantial advances have been made in understanding the molecular basis and clinical manifestations of cardiomyopathy, however little is known about why disease expression and outcomes are so variable – manifesting as age of onset, severity of remodelling, and the cumulative lifetime burden of cardiovascular events. We are exploring whether task-driven clustering of high-dimensional data can optimally define cardiomyopathy classes by outcome and response to therapy compared to conventional risk groups. We are also attempting to understand the transition from health to disease by determining if pathogenic variants in cardiomyopathy-associated genes predispose to phenotypic adaptations in the general population and relatives of probands.
Identifying disease-modifying molecular pathways: We are investigating whether common polymorphisms and low-frequency missense variants in genes of interest contribute to a genetic predisposition to altered mechanical and tissue characteristics – using large-scale imaging and whole-exome sequencing in UK Biobank. These approaches harness the power of machine learning for high-throughput imaging-genetics research on large populations to prioritise genes for which pharmacologic agents may lower risk for disease.
Software – software and other tools developed by the group are available at https://github.com/UK-Digital-Heart-Project.





McGurk KA, Qiao M, Zheng SL, Sau A, Henry A, Ribeiro ALP, Ribeiro AH, Ng FS, Lumbers RT, Bai W, Ware JS, O’Regan DP. Genetic and phenotypic architecture of human myocardial trabeculation. Nat Cardiovasc Res. 2024. doi: 10.1038/s44161-024-00564-3
Curran L, de Marvao A, Inglese P, McGurk KA, Schiratti PR, Clement A, Zheng SL, Li S, Pua CJ, Shah M, Jafari M, Theotokis P, Buchan RJ, Jurgens SJ, Raphael CE, Baksi AJ, Pantazis A, Halliday BP, Pennell DJ, Bai W, Chin CWL, Tadros R, Bezzina CR, Watkins H, Cook SA, Prasad SK, Ware JS, O’Regan DP. Genotype-Phenotype Taxonomy of Hypertrophic Cardiomyopathy. Circ Genom Precis Med. 2023;16(6):e004200. doi: 10.1161/CIRCGEN.123.004200
Shah M, de A Inácio MH, Lu C, Schiratti PR, Zheng SL, Clement A, de Marvao A, Bai W, King AP, Ware JS, Wilkins MR, Mielke J, Elci E, Kryukov I, McGurk KA, Bender C, Freitag DF, O’Regan DP. Environmental and genetic predictors of human cardiovascular ageing. Nat Commun. 2023;14(1):4941. doi: 10.1038/s41467-023-40566-6
Thanaj, M., Mielke, J., McGurk, K.A., Bai W., Savioli N., de Marvao A., V Meyer H., Zeng L., Sohler F., Lumbers R. T., Wilkins M. R., Ware J. S., Bender C., Rueckert D., MacNamara A., Freitag D. F., O’Regan D. P. (2022). Genetic and environmental determinants of diastolic heart function. Nat Cardiovasc Res. 1, 361–371. https://doi.org/10.1038/s44161-022-00048-2
Meyer HV, Dawes TJW, Serrani M, Bai W, Tokarczuk P, Cai J, de Marvao A, Henry A, Lumbers RT, Gierten J, Thumberger T, Wittbrodt J, Ware JS, Rueckert D, Matthews PM, Prasad SK, Costantino ML, Cook SA, Birney E, O’Regan DP. (2020). Genetic and functional insights into the fractal structure of the heart. Nature. 584; 589–594.
Bai W, Suzuki H, Huang J, Francis C, Wang S, Tarroni G, Guitton F, Aung N, Fung K, Petersen SE, Piechnik SK, Neubauer S, Evangelou E, Dehghan A, O’Regan DP, Wilkins MR, Guo Y, Matthews PM, Rueckert D. (2020). A population-based phenome-wide association study of cardiac and aortic structure and function. Nature Medicine. https://doi.org/10.1038/s41591-020-1009-y.
Bello GA, Dawes TJW, Duan J, Biffi C, de Marvao A, Howard LSGE, Gibbs SR, Wilkins MR, Cook SA, Rueckert D, O’Regan DP. (2019). Deep learning cardiac motion analysis for human survival prediction. Nature Machine Intelligence. Vol:1, ISSN:2522-5839; 95-104