By Sophie Arthur
September 1, 2020
Time to read: 6 minutes
The sequence of DNA is the fundamental blueprint to build a complex organism like a human. All the different organs in the body, each made up of millions of individual cells, are descended from one single cell and as a result, they have exactly the same sequence of DNA. Yet the different types of cells in the body have very different functions. This is possible because different cells use different parts of the DNA sequence. How does this work?
Imagine you have several copies of the same book, with the same text. Then imagine that you take one copy of the book and use a marker pen to cross out all of the text in one chapter. In a different copy, you do the same thing but for a different chapter. Then, the result of reading the book will be different in each copy. This is similar to what happens when a body develops from a single cell. For example, cells in the pancreas make the hormone insulin from the insulin gene in the DNA. The gene for insulin is therefore turned on in pancreatic cells. However, cells in the liver shut down the insulin gene permanently and don’t “read” it – the gene is turned off. So, there must be an extra level of information beyond the DNA sequence, which is required to build an organism from a single cell. This is known as epigenetic information.
An important feature of epigenetic information is that it is inherited, just like the DNA itself, when cells divide. This process is very accurate. It has to be otherwise cells that divide would forget what they are supposed to be doing, with potentially disastrous consequences. As a result, there are mechanisms that propagate epigenetic information through divisions of cells. Amazingly, sometimes these mechanisms are so strong that epigenetic information can be passed on between generations – so-called “transgenerational” epigenetic inheritance. This is surprising and interesting for many different reasons and many researchers are studying why it happens. But my laboratory has been studying one particular question raised by the existence of transgenerational epigenetic inheritance: can it contribute to evolution?
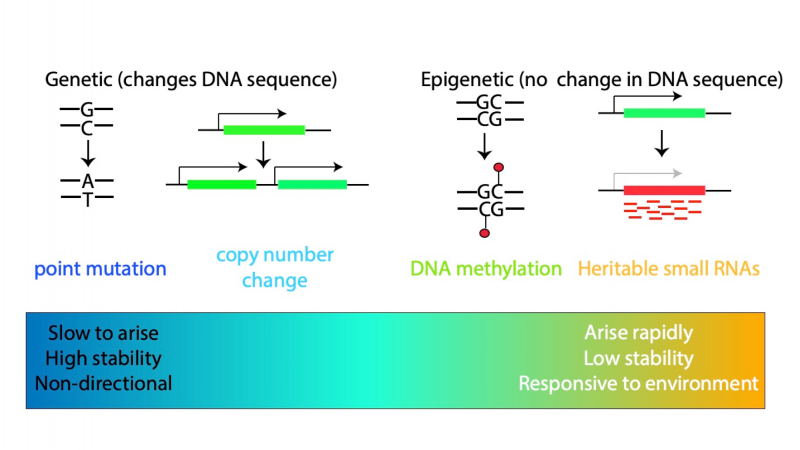
In the traditional view of evolution, changes that occur in species over long periods of time are caused by changes in the DNA sequence, known as mutations. These can alter how the organism develops. Some of these changes might be beneficial, making the organism able to survive better or have more offspring, and over time these are more likely to become common in the species. We wondered whether epigenetic changes, which don’t alter the DNA sequence but instead change the way genes are used, could behave in the same way as changes in DNA.
To go back to the book analogy, imagine you gave the two different copies, with two different chapters obliterated, to different people to copy. If they copy only the text, then the original book you started with all the chapters would be recovered by both. However, if they copy the marker pen as well, the copies would be different when read, without having any differences in their texts. We term these epigenetic changes as “epimutations” to contrast them with mutations in the DNA sequence.
We wanted to know if epimutations could contribute to evolutionary processes. For this to be possible the epimutations would have to last for many generations, just like a DNA sequence change which, once it has happened, is very unlikely to disappear. To study this question, we needed to use an animal that we could grow for many generations in the lab in a reasonable time! So, we decided to use a simple worm, called C. elegans, for our study. An additional advantage of C. elegans for this particular experiment is that the process of epigenetic silencing has already been well studied. In C. elegans, some artificial genes inserted into the genome can be turned off by creation of “small RNAs” – pieces of RNA about 20 nucleotides in length which are able to bind to the mRNA of genes and signal for them to be shut down. We were therefore able to investigate whether some of the genes in C. elegans were stably shut down by small RNAs and, if so, how long this lasted for.
To test these ideas, in collaboration with the lab of Vaishali Katju in Texas, we grew 10 independent populations of C. elegans, all descended from the same ancestor, for up to 100 generations. We investigated their small RNAs to detect epimutations at 25 generations and at 100 generations, and compared to the ancestral population. We found many differences, about 100 genes in each line after 25 generations, suggesting that epimutations did occur quite frequently. For comparison, only 1 DNA sequence change on average occurred in each line over this time.
However, if we compared the same line at 25 and 100 generations, very few of the changes were the same! This meant that either the changes were happening randomly every generation (so not epimutations at all), or that epimutations were happening but were disappearing again quickly and being replaced by different epimutations.
To understand what was going on we needed to do a more detailed time course, where we investigated epimutations in populations every generation, for up to 13 generations. Toni Beltran, a PhD student in the lab, performed this heroic experiment, which involved coming in to the lab at strange times of day and night to collect worms. His immense dedication paid off because with this data we were able to precisely map how long an epimutation lasted for on average: around 3 generations! So, this explained exactly why the epimutations at 25 generations had disappeared by 100 generations: they cannot be maintained for this length of time. We now know for the first time how stable epimutations are in an animal.
What is the significance of this observation for evolution? It’s clear that epimutations cannot drive evolutionary processes over 100s of generations because they do not last long enough. However, we think that they might have a role in the very early stages of the process of evolution, perhaps by allowing a few animals to survive a stressful environment. Epimutations occur much more rapidly than DNA sequence changes so this would give time for a DNA sequence change to arise which would then cement the adaptation. We are now trying to test this idea in the laboratory – it’s very exciting as it might change the way we think about evolution, but at the moment it’s still only a hypothesis!
‘Epimutations driven by small RNAs arise frequently but most have limited duration in Caenorhabditis elegans’ was published on 31 August in Nature Ecology and Evolution. Read the full article here.
Written by Peter Sarkies, Head of the Epigenetic Inheritance and Evolution group at the MRC LMS.