By Sophie Arthur
July 17, 2020
Time to read: 4 minutes
DNA replication is a critical process that our cells must perform. It is essential for growth, genome stability, inheritance and ageing. A key step in this process is the formation of a so-called replication fork. This is where the DNA helix becomes unzipped into two single strands, which is performed by an enzyme called a helicase. In order for the helicase to unwind the DNA, it must first be loaded onto the DNA strand. But the structural changes that a helicase must go through to do this have been a mystery for many years.
Research by the DNA Replication group at the Institute of Clinical Sciences and the MRC LMS has now revealed this mechanism of helicase loading. The group used a technique called cryo-electron microscopy (cryo-EM) and computational modelling, described in a paper published in Proceedings of the National Academy of Science in the USA (PNAS) on 15 July 2020. The researchers investigated the structural changes in the core of the helicase, which is made up of six minichromosome-maintenance 2-7 (Mcm2-7) proteins. They also revealed how the core Mcm2-7 proteins dock with the helicase loader so that DNA can be inserted into the helicase core – the central process of helicase loading.
Intriguingly, the team found that small genetic modifications in these Mcm2-7 proteins slowed down the process of helicase loading. The reduced tempo allowed the researchers to study the intermediate structures in loading in great detail. They found that the helicase loading process has four key steps; initial anchoring to the loader, docking of the helicase core, DNA insertion and closing of the Mcm2-7 core.
The first structure shows part of the Mcm2-7 proteins, called the C-terminal domain, reaching out to make initial contact with the helicase loader, like antennae reaching out to examine the surrounding environment. In this first step, only two of the six proteins in the helicase core bind to the helicase loading complex. By the end of step two, all Mcm proteins in the helicase core are bound to the helicase loader.
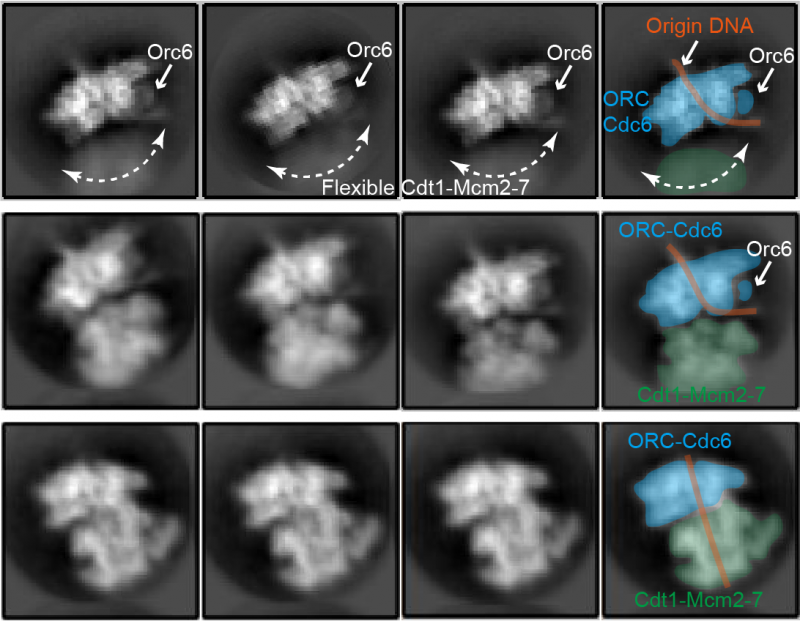
The third step is the crucial DNA insertion, where key structural changes cause the Mcm proteins in the helicase to form a ring around the DNA strand. In a switch from cryo-EM, the computational modelling approach revealed the structural changes which have to take place for DNA insertion to occur. In fact, the observed Mcm2-7 ring has to widen.
The final step is for the Mcm protein ring to completely close, encapsulating the DNA in preparation for the initial steps of DNA replication. Computational models revealed that electrostatic charges on the DNA – positive or negative charges generated by friction between two surfaces – pull the Mcm2-7 proteins around it. This creates a self-sealing structure where the helicase is loaded onto the DNA.
This work was performed in yeast, but as the helicase loading system is highly conserved from yeast all the way to humans, the four-step process described in this study is likely to apply in people too.
Prof Christian Speck, Head of the DNA Replication group at Imperial College London, who is based at the MRC LMS and senior author on this study, discussed its possible implications for healthcare:
“There is a need to look for novel inhibitors of the DNA replication process. One particular example is for chemotherapy, which is very efficient at killing tumour cells, but also generate lots of mutations that can lead to relapse. So, blocking DNA replication at an early stage may be one way to kill tumour cells without generating mutations. Inhibitors that block any of these steps, and prevent DNA replication, will be the next step for our research – in collaboration with Alexis Barr of the LMS’ Cell Cycle Control group and the Department of Chemistry at Imperial College London. I also want to acknowledge the incredible work that all my colleagues have contributed to this study, but particularly Sarah Schneider.”
‘Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6’ was published on 15 July in PNAS. Read the full article here.