By Helen Figueira
January 2, 2014
Time to read: 4 minutes
 How computational models can advance medical research
How computational models can advance medical research
It’s hard to think of a time before the Internet. Before Facebook and Twitter governed our social lives, LinkedIn kept our professional lives in check, and the likes of Amazon and Netflix guided our next book or movie purchase. Networks drive today’s society. And now, new research from the CSC uncovers how gene networks can push forward our understanding and treatment of complex diseases.
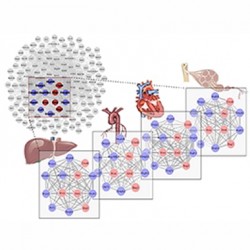
Enrico Petretto’s team (Integrative Genomics Medicine Group) devised a new, flexible and stable computational method that can analyse large-scale genomic data, across multiple systems, and pool together networks that allow exploration into the intricate interactions that govern biological processes. “Over the last few years, a vast stream of genomic data has risen exponentially and are made available to the scientific community,” explains Enrico, “investigators can download these data and analyse accordingly to their specific biological questions. However, the main limitation is in the ability of accessing only specific ‘slices’ of the data; for example, protein-protein interaction within a single tissue, or a single disease, and as such inevitably miss out on the bigger picture.” Enrico’s method, published in PLOS Genetics, overcomes such limitation by allowing multiple genome-scale datasets to be analysed collectively in a single step, at the systems level.
By testing their model on real-life datasets, Enrico’s team identified two new biological findings. “We picked up a network of genes, co-expressed in multiple tissues in rats, which link Heat shock protein (HSP) to dilated cardiomyopathy – a condition in which the heart becomes weakened and enlarged.” While such a link has up until now only been speculated, Enrico’s team were the first to show this using their new computational approach. “Because we could analyse gene expression patterns simultaneously in seven rat tissues, we were able to identify and highlight such a network.” Enrico’s team next applied their model to a completely different dataset – gene expression in human developing brain. They were interested in which brain area and when, during brain development, certain pathways (e.g., calcium signalling, axon guidance) became functionally active. Previous work suggests these pathways, central to cognitive function, only became functional much later during brain development. While, Enrico’s team revealed the opposite: that the pathways are already active early on in the developing brain.
Short-term, the method’s labour-saving characteristic sets to benefit biologists significantly in their exploratory analysis. “Let’s say you have a new dataset that you want to explore at the systems-level, you can render our approach and generate a hypothesis; then test the in silico generated hypothesis in the lab.” Longer-term, the outcome would be a better understanding of the networks and pathways of complex diseases that would guide informed treatment approaches. “We have to move to systems-level analyses. You could have 100 genes that are contributing to a disease, unless you look at all of them, and in different biological contexts (e.g., different tissues), you won’t know which is the best one for therapeutic target,” adds Enrico.
Facebook, Twitter and LinkedIn succeed through allowing connections and interactions to be made through a host of networks. Enrico’s team share the same philosophy: viewing genetic systems as a whole, rather than gene-by-gene, to form large-scale networks. And with genome projects such as 23andMe and Genomics England in full swing, there is no shortage of data. What’s more, methods that use the genomic data boom to their advantage – as fuel even – will only ensure all possible key players – whether previously known or not – are considered in therapeutic strategies.
YJ
Reference: Xiao, X., Moreno-Moral, A., Rotival, M., Bottolo, L., & Petretto, E. (2014). Multi-tissue analysis of co-expression networks by Higher-Order generalized singular value decomposition identifies functionally coherent transcriptional modules. PLoS Genetics, 10 (1), e1004006.
This work was supported by the European Community’s Seventh Framework Programme, the Medical Research Council, the British Heart Foundation and from EPSRC Mathematics Platform.
The Medical Research Council has been at the forefront of scientific discovery to improve human health. Founded in 1913 to tackle tuberculosis, the MRC now invests taxpayers’ money in some of the best medical research in the world across every area of health. Twenty-nine MRC-funded researchers have won Nobel prizes in a wide range of disciplines, and MRC scientists have been behind such diverse discoveries as vitamins, the structure of DNA and the link between smoking and cancer, as well as achievements such as pioneering the use of randomised controlled trials, the invention of MRI scanning, and the development of a group of antibodies used in the making of some of the most successful drugs ever developed. Today, MRC-funded scientists tackle some of the greatest health problems facing humanity in the 21st century, from the rising tide of chronic diseases associated with ageing to the threats posed by rapidly mutating micro-organisms. www.mrc.ac.uk