By Lindsay Keith
November 17, 2022
Time to read: 3 minutes
A groundbreaking mathematical model which helps us to understand what factors affect the speed at which cells replicate has been developed by researchers from the MRC London Institute of Medical Sciences (LMS) and Imperial College London. Using single-cell live imaging data, they investigated cell-cycle factors that could affect the interdivision time of cells – that is the length of time taken for one cell to replicate and generate two new cells.
Cell replication is a vital part of healthy human physiology but, when it becomes dysregulated or excessive it can cause or contribute to a number of diseases. The excess replication of cells is known to be a factor in the development of cancer, atherosclerosis, and rheumatoid arthritis, amongst others. Fully understanding the factors that control cell replication is therefore vital towards developing treatments for such diseases.
Cell cycle factors are hidden properties, i.e., they are not visible in the data, but they influence interdivision times. The challenge is to try and determine the nature of these factors. In their most recent publication, the Cell Cycle Control group led by Dr Alexis Barr, in collaboration with Dr Philipp Thomas in the Department of Mathematics at Imperial College London, propose a model able to predict these hidden cell cycle factor dynamics using the correlation patterns of interdivision times.
“Our proposed framework of inheritance of interdivision time from one cellular generation to the next allows us to interpret factors that affect the length of the cell cycle in a range of cell types. We often find some hidden rhythmicity in this inheritance.” – Fern Hughes
It is currently unclear how interdivision time patterns can provide information about the cell cycle factors. Using single-cell imaging data, the group was able to look at interdivision times with a lineage (family) tree structure. This analysis then uses these trees to investigate how the interdivision time of a cell is related to that of others in the tree – such as its mother cell, sister cell or cousin cell. This in-depth exploration of these correlations reveals several characteristics of cell cycle factors hidden in lineage tree interdivision time data.
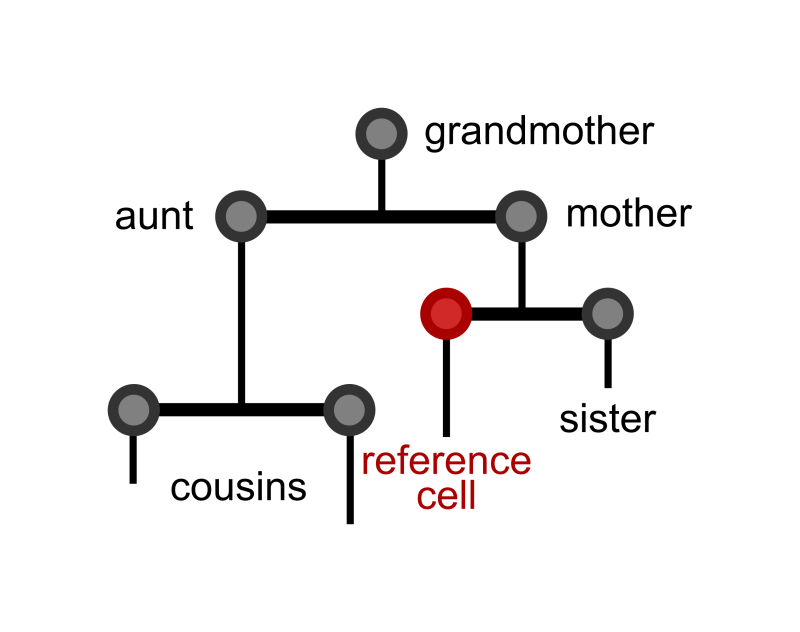
“Previous work using single-cell imaging data has revealed non-intuitive cell cycle length correlations between cells that share a common ancestor. In this work we propose a mathematical model to decode these correlation patterns and reveal the factors that affect the length of the cell cycle” explains Fern Hughes, the lead author of the paper.
Fitting the single-cell imaging data, the model can predict the factors that affect the cell cycle length. Specifically, a probabilistic approach was used, explains Fern Hughes:
“The parameters are fit using Bayesian approach, meaning that our predictions are presented as distributions, allowing us to assess the probability of each outcome.”
This model is applicable to many different cell types and allows for a non-invasive way to study cell cycle factors, just by measuring interdivision lengths across generations of cells. This model also has the potential of exploring the factors that affect other measurable quantities besides cell cycle length.
“We take a data-driven approach to avoid any prior assumptions on the cell division mechanisms, allowing the data to speak for itself.” – Fern Hughes
“This work makes an important contribution to the study of the cell cycle”
– Editors from eLife.
This research was published in eLife and was led by Alexis R. Barr and Philipp Thomas. This research was conducted by researchers at the LMS in collaboration with Imperial College London and was funded by the MRC-LMS and the EPSRC Centre for Precision Healthcare.
This news article was written by Isabella Giokari