By Sophie Arthur
November 8, 2020
Time to read: 6 minutes
By Dr Peter Sarkies
Very few scientific publications are announced simultaneously by the UK Prime Minster and the US President. Twenty years ago in June 2000, Tony Blair and Bill Clinton announced the publication of the draft sequence of the human genome, immediately catapulting the field of genomics into the public domain. Twenty years later, and we are still grappling with the implications of knowing the exact order of the 3 billion instances of the 4 DNA bases (ACGT) that make up our genome. At the time, the transformative insights revealed were relatively few. There was one point, however, that stood out immediately from the string of letters.
A gene is an individual unit that specifies the production of a single molecular actor, often a protein but sometimes an RNA molecule, that helps in the construction or activity of a cell. Together, the genes in a genome specify how an organism develops and functions. The genome of an organism was thus considered to be a sequence containing the genes, perhaps with some segments of DNA that helped to control where and when these genes would be active. Indeed, some of the first genomes sequenced, such as the bacteria E. coli and the yeast S. cerevisiae, confirmed this view. But it was also known that the human genome was much bigger than either of these, and so scientists mostly assumed that the human genome contained many more genes, as we are much more complex organisms. It all seemed to fit very nicely. However, the publication of the human genome sequence revealed immediately that this was not at all true. The human genome doesn’t contain many more genes than yeast. Instead, the massive size of the human genome reflects the fact that 99% of it doesn’t code for genes at all!
What does this silent material do? One of the most puzzling constituents of the non-coding portion of the genome are so-called “Transposable Elements”, known as TEs for short, which total about 40 per cent of the genome. Transposable elements are a bit like viruses that have infected the genome, rather than cells. They can copy themselves independently of the rest of the genome and insert these copies elsewhere. Over time, the number of TEs grows and the genome expands as a result. However, the host organism fights back using a variety of mechanisms to shut down TEs to stop them taking over. My lab has been studying these mechanisms, and has published several papers about how these defence mechanisms evolve across species.
It is still unclear why some genomes, like ours, are so full of TEs, whereas others are not. The yeast genome, for example, has hardly any at all. Others, like pine tree genomes, are comprised of up to 80 per cent TEs. This has led scientists to speculate whether TEs confer some kind of hidden benefit, which might explain why humans keep them in the genome rather than destroying them completely. My lab became interested in this question, and to answer it we decided that we needed to watch genomes evolve over many generations in the lab.
To do this we collaborated with a lab at Texas A&M University in the USA, run by Vaishali Katju. We used a simple worm, known as C. elegans, which has a three-day lifecycle and can be propagated for many hundreds of generations. Crucially, we grew the worms at three different population sizes. Either one individual, 10 individuals or 100 individuals were selected each generation and allowed to replicate. This is very important because it allows us to investigate the effect of natural selection.
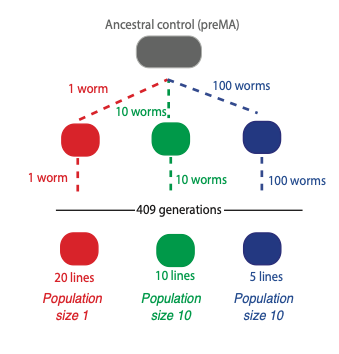
Natural selection is very ineffective when only one individual is selected in each generation, because there is no competition within the population and so all of the changes that occur in the one animal are passed on, whether they are good or bad. With a larger population size, such as 100 worms, bad changes can be eliminated by natural selection so only good or neutral changes are passed on. By comparing small and large populations we can see which changes are detrimental, because these will occur in small population sizes but not in large ones.
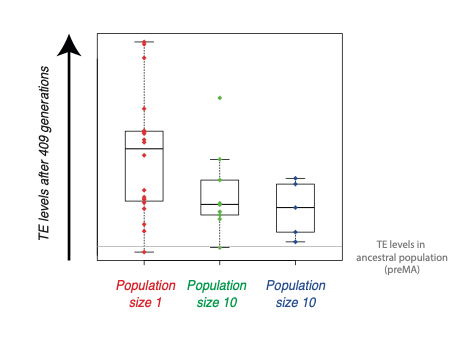
Together with Professor Katju, we used this setup to investigate whether TEs are generally beneficial, neutral or deleterious. We examined the levels of TEs after more than 400 generations of propagation in the lab, and compared different population sizes. The result was very clear: the levels of TEs were much higher in populations with small sizes of one individual than in those containing 10 or 100 individuals. This indicates that in general, TEs are likely to be detrimental.
What intrigued me about this result was the question of how the levels of TEs were increasing. One possibility was that the mechanisms that the cells use to shut down TEs were becoming less effective over time in the small populations. We decided to test this by looking at silencing of TEs by a type of RNA known as a Piwi-interacting RNA, or piRNA for short. Cells make piRNAs to search for TEs and shut them down. piRNAs are able to do this because they have similar sequences to parts of the TEs. Remarkably, we discovered that in population sizes of one individual, piRNA silencing of certain TEs was failing, and this is why TE levels were increasing.
What does this mean for understanding human genomes? Our results suggest that, in general, expanding levels of TEs are not good for organisms. Their levels are a delicate balance between the drive of the TE to copy itself and the power of the molecular mechanisms that the cell puts in place to stop them. Of course, the remaining question is what causes this balance to be different in different organisms – why are there so few TEs in yeast and so many in humans? We’re hoping that further experiments we are doing to evolve worms in the lab might answer these questions.
‘Long-term experimental evolution reveals purifying selection on piRNA-mediated control of transposable element expression’ was published on 6 November in BMC Biology. Read the full article here.