By Sophie Arthur
February 8, 2019
Time to read: 5 minutes
Have you ever been watching an orchestral performance and tried to pick out the tune that the trombonists are playing? It is a challenge to pick out that one melody that you are trying to focus on. Of course, you might be able to pick up short snippets of the iconic riff the trombones are playing, but maybe you’re not getting the whole story.
The same goes for a population of cells. Quite often, any differences in the expression of genes or proteins from an experiment is showing us the average change in that population. But that average effect that is reported is actually hiding the underlying changes in each individual cell of that population. Surely, knowing the amount of difference, or heterogeneity, within that cell population could uncover more truths about what really goes on in an individual cell and how they respond and adapt.
Research published in Nature Microbiology on 4 February from the Quantitative Gene Expression group at the LMS used single celled organisms, in this case yeast, to reveal how different cells really are and how they are influenced by their environment.
In this study the researchers used a dual approach to explore how diverse the individual cells of yeast were. They were able to acquire a picture of each individual cell, break open the cell walls, extract the RNA and amplify it enough to sequence the transcriptomes. This was complemented by a theoretical approach developed by researchers from the Department of Mathematics at Imperial College London. By using Bayesian statistics, the researchers were able to make sense of the very noisy yeast data.
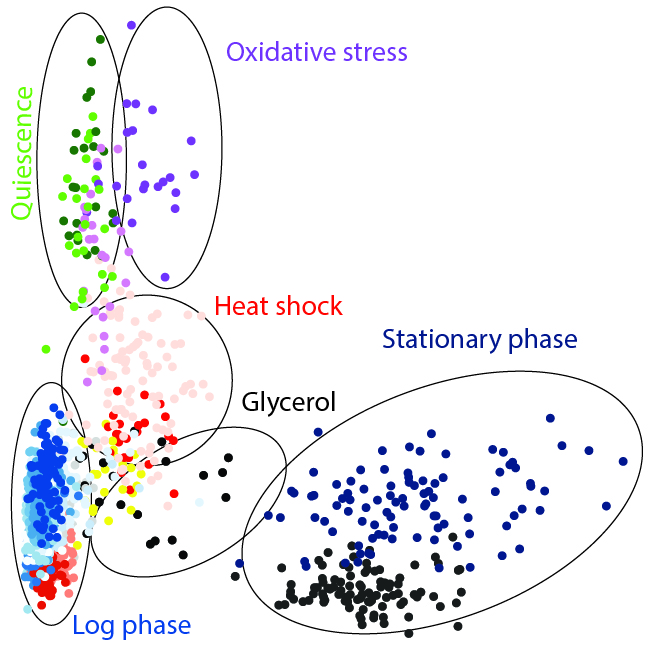
The findings in this study revealed that when the yeast cells were growing happily there was very little variability amongst the cells. The main source of variability was at which stage of the cell cycle each cell was in. But as the cells began to grow in media progressively exhausted of its nutrients, the change in environment induced a gradual stress response in the cells in synchrony. That was until a critical point when a very strong transcriptional heterogeneity appeared. Interestingly, when the researchers changed the cells’ environment through a strong and sudden heat shock, the cells responded again all in the same way.
Learning about the impact of environmental changes on how each cell in a population responds could potentially change how drug treatments work. Further research is required but these findings suggest that a stressful environment could be making individual cells more or less resistant to the drugs by inducing gene expression heterogeneity. Therefore, understanding the heterogeneity could help us to kill more cells by learning how certain cells escape treatments such as antibiotics or chemotherapy.
This work was part of an interdisciplinary collaboration with Vahid Shahrezaei, Department of Mathematics at Imperial College London. Samuel Marguerat, Head of the Quantitative Gene Expression group at the LMS and senior author of the paper told us about the importance of the interdisciplinary nature of this work:
“Working with single cell RNA data is tricky because for a lot of genes you get a reading of zero transcripts. This mathematical approach allowed us to clean up the data and undercover whether that zero was because that gene isn’t expressed or because of some technical issues. It has allowed us to make more sense of the data amongst all the noise.”
Some genes are known to be regulated with cell size, and by knowing cell size and the variability allows better prediction of gene expression. Gene expression is often thought to be random, but by measuring more variables in differing environments, we are exposing that it may be less random than it appears.
The next steps for this work are to further understand the roots of the heterogeneity, and what types of stress or duration of environmental change cause these differences before adding layers to this technology with more cells, more genetic information and more protein information to build a bigger and more complete picture. Adding more parts around the trombonist’s melody again to create a beautiful symphony where we understand how all the layers fit together.
Samuel Marguerat wrote a blog piece for Nature Microbiology about this work which you can read here.