By Jenna Stevens-Smith
October 12, 2018
Time to read: 4 minutes
In every cell of our body gene machines are able to turn genes on/off and flag damaged areas of DNA for repair. This is possible due to enzymes known as chromatin remodellers, which do this by constantly unwrapping and wrapping the DNA in our cells.
Research published in Science on 11 October provides new insight to the function of one of these chromatin remodellers, SWR1, known to flag areas of damaged DNA, which if not flagged could lead to cancer in humans.
The research is a collaboration between LMS and Imperial researchers David Rueda, head of the Single Molecule Imaging group and Chair of Molecular and Cellular Medicine, and Dale Wigley, Chair in Protein Crystallography in the Department of Medicine. The team used their complementary techniques of cryo-Electron Microscopy (cryo-EM), to solve the structure of SWR1, and single molecule imaging, to visualise how SWR1 unwraps the DNA. Through this combined approach the researchers gained new insight to the mechanism, helping to improve our understanding of how cells repair damage to their DNA. The researchers used yeast as the model organism in this study.
How does it work?
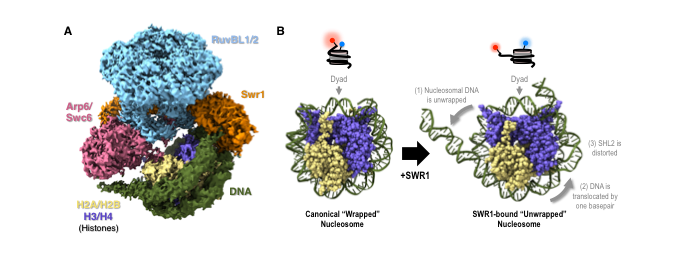
Every cell in our bodies has the equivalent of 2m of DNA packed inside its microscopic nucleus. To pack this much DNA into each cell, the DNA is wrapped around a complex of proteins called histones. The DNA wrapped around the histones is known as nucleosomes, and the strands of the packaged DNA referred to as chromatin. When DNA is tightly wrapped around the histones for packaging into the nucleus of a cell it is unable to express genes. The function of SWR1 is to drive the exchange of one of the histone protein pairs H2A:H2B for H2AZ:H2B. It does this by initially unwrapping a small part (~10-12 base pairs) of the nucleosome DNA from around the histone complex. The exchanged histone protein, H2AZ (or Htzl), acts as the flag for the DNA repair machinery within the cells.
The role of ATP
Through the structural and single molecule imaging techniques the researchers identified that the histone exchange is only possible if ATP binding occurs. This was due to a slight, but essential, distortion of the DNA wrapped around the histone proteins, caused by the ATP binding. The binding causes the distortion of the structure, which is essential for the unwrapping of the DNA to occur. Similarly, if the SWR1 enzyme is not present the histone exchange will not occur even if there is ATP binding.
Start and stopping with cryo-EM
Cryo-EM is able to provide high-resolution structural information about biomolecules such as proteins and nucleic acids in solution. In this project, the cryo-EM team, led by Dr Wigley were able to identify the start (wrapped) and finish (unwrapped) structure. However, the cryo-EM was not able to provide information about the intermediate steps between the wrapped and unwrapped states.
Lighting up the unwrapping of DNA
Single molecule imaging was used to understand the dynamics of the intermediate steps of the unwrapping of the DNA. To do thus the single molecule imaging team, led by Dr Rueda, fluorescently tagged two parts of the DNA wrapped around the histone. Depending on the proximity of these tagged areas to each other there was either a red fluorescence detected (wrapped DNA- high ratio) or a green fluorescence detected (unwrapped DNA- low ratio).
David Rueda, head of the Single Molecule Imaging group at the LMS and joint senior author for the study said:
“The combined approach of structural and dynamic imaging in this publication is promising. Through these collaborations we will gain a better understanding of how chromatin remodellers such as SWR1 are able to flag damaged areas of DNA for repair. It is important to understand how these processes function and how they may go wrong in disease. This understanding could lead to new ways to prevent or treat cancer and other diseases in the future.”
The full publication, Structure and dynamics of the yeast SWR1-nucleosome complex, can be read here.
Oliver Willhoft, Mohamed Ghoneim, Chia-Liang Lin, Eugene Y. D. Chua, Martin Wilkinson, Yuriy Chaban, Rafael Ayala, Elizabeth A. McCormack, Lorraine Ocloo, David S. Rueda and Dale B. Wigley (2018) Structure and dynamics of the yeast SWR1-nucleosome complex Science 362 (6411), eaat7716. DOI: 10.1126/science.aat7716