By Jenna Stevens-Smith
November 2, 2018
Time to read: 5 minutes
Researchers have gained insight into how an essential protein for cell function regulates gene activity, which may serve as a future target for cancer treatment.
The research, published in Molecular Cell on 1 November, provides novel understanding of the steps the protein ubiquitin/UHRF1 plays in determining protein breakdown and growth in cells.
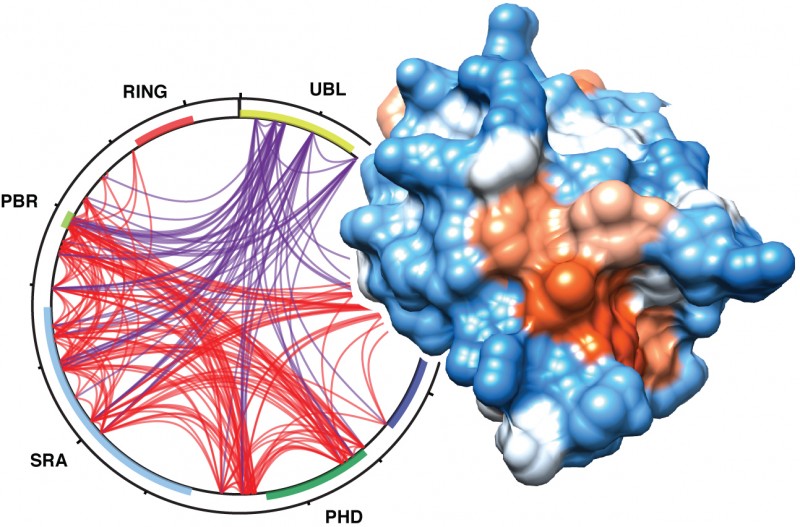
It highlights how UHRF1 regulates chromatin, the complex of DNA and proteins which enables DNA to be packaged into the nucleus of cells. The findings indicate the importance of UHRF1 in the process of gene expression regulation. It is known that control of gene expression is also fundamental in the prevention and treatment of cancer.
In order to begin to think about potential treatments, researchers need to understand how the potential target protein works. The key finding of this research is that UHRF1 catalyses a critical enzymatic step in the marking of methyl groups on DNA prior to DNA methylation itself, a process of epigenetic control of gene expression.
The critical step is the ubiquitylation, which is the process of the final attachment of ubiquitin to a protein, in this casehistone H3. Histones are complexes of proteins that DNA wraps around forming chromatin, in order to be packed into the nucleus of a cell.
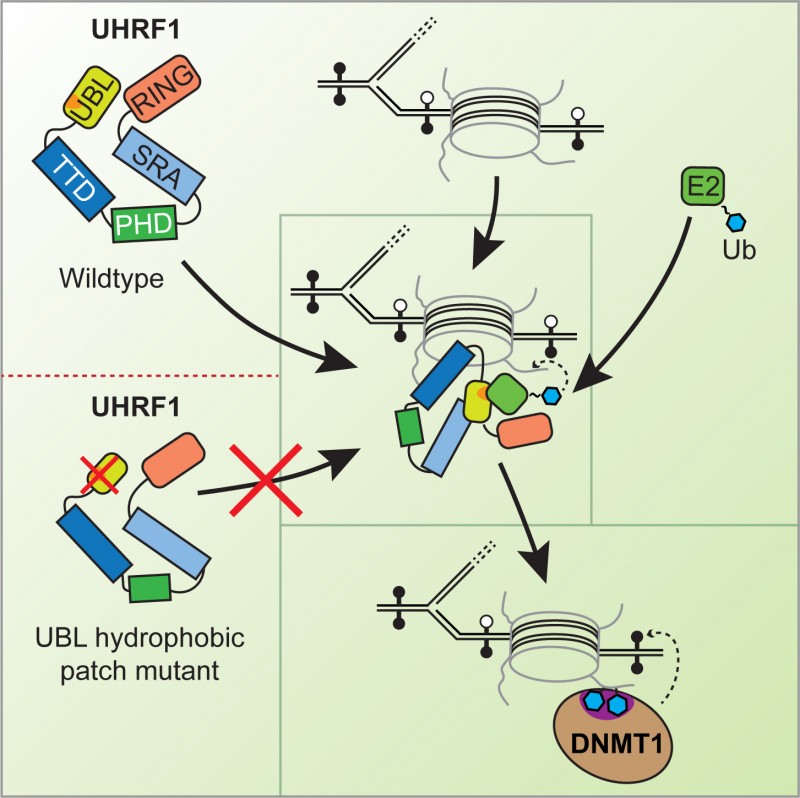
Without ubiquitylation DNA methylation cannot happen. Without DNA methylation normally silenced regions of the genome become activated leading to genomic instability. By understanding the enzymatic mechanism of how UHRF1 ubiquitylates histone H3 in chromatin the researchers have shown that interfering with the mechanism leads to defective DNA methylation. The key physical characteristic of the UHRF1enzyme complexthat they identified, is the Ubiquitin-like domain (UBL) which mediates the ubiquitylation of histone H3. This region is an essential requirement for the complete transfer of ubiquitin to chromatin by UHRF1 and explains why chromatin could not be ubiquitylated without UHRF1.
This research shows how complex the biochemical and structural interactions involved in the transfer of the signalling protein ubiquitin are, but also the many mechanisms in place to ensure this takes place without error.
It was a collaboration between the Institute of Functional Epigeneticsat Helmholtz Zentrum München, the MRC LMS and Ludwig Maximilians University Munich (LMU). The MRC LMS Proteomics Facility performed analysis using a cross-linking mass spectrometry approach. Sebastian Bultmann at the LMU was able to show, using CRISPR/Cas9 knock-ins, that the UBL domain is essential for DNA methylation in cells.
Holger Kramer, Head of the Proteomics Facility at the LMS, shared the use of the cross-linking approach within this study and the interests of the Proteomics Facility,
“This is the first project where we have exploited the cross-linking approach and we, along with research labs in the Institute, are interested in further exploring this type of technology and utilising these approaches to complement our work. As a facility one of the things we are really interested in is the general field of protein modifications and that in itself is so big. I think that the fascinating thing with proteomics and protein biochemistry is that there is so much detailed information about this level of biological control that can be accessed.”
Till Bartke, Deputy Director, Institute of Functional Epigenetics, and senior author of the paper, shared his praise for first author, Ben Foster and the next steps for the research,
“Ben Foster the first author has single-handedly conceived the project and designed and carried out all the biochemical experiments. He has done a tremendous amount of work and he deserves all the credit. We had a very productive collaboration with Alex Montoya and Holger Kramer from the proteomics facility at the MRC LMS. They were very helpful and proactive in establishing the chemical cross-linking and mass-spectrometry protocols together with Ben.
The next steps will be to do more detailed biochemical studies with tailored modified chromatin substrates to understand how UHRF1 directs the ubiquitylation to the correct site on histone H3.”
The study sheds light on UHRF1’s enzymatic mechanism and indicates possible ways to interfere with its function. This knowledge could be used for developing drugs for future cancer therapies. To find out more about the research read the full paper, Critical role of the UBL-domain in stimulating the E3 ubiquitin ligase activity of UHRF1 towards chromatin.
Further Information
Foster, B. M. et al. (2018): Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatin. Molecular Cell, DOI: 10.1016/j.molcel.2018.09.028
Dr Till Bartke is the Deputy Director of the Institute of Functional Epigenetics (IFE) at Helmholtz Zentrum München – German Research Center for Environmental Health. Prior to joining IFE in 2017 Till was a Group Head at the MRC London Institute of Medical Sciences.
This article was written by Angel Thomas, Scout Davies and Jenna Stevens-Smith.