By Staff Member
March 2, 2018
Time to read: 3 minutes
RNA molecules play many important roles in fundamental cellular processes such as control of gene expression and protein synthesis, they may even have initiated life on earth. But imaging these molecules, to understand their functions in their physiological environment, has been a major challenge.
An MRC funded team of researchers, led by Professor David Rueda, Head of Single Molecule Imaging at LMS, have just reported a new set of tools to image cellular RNAs in live and fixed cells. These tools hold immense potential to surpass existing tools and were developed and tested as part of an international collaboration between MRC LMS, a Canadian team led by Dr. Peter Unrau, Professor of Molecular Biology and Biochemistry at Simon Fraser University; and a French team led by Dr Michael Ryckelynck, Head of Digital Biology of RNA at the University of Strasbourg.
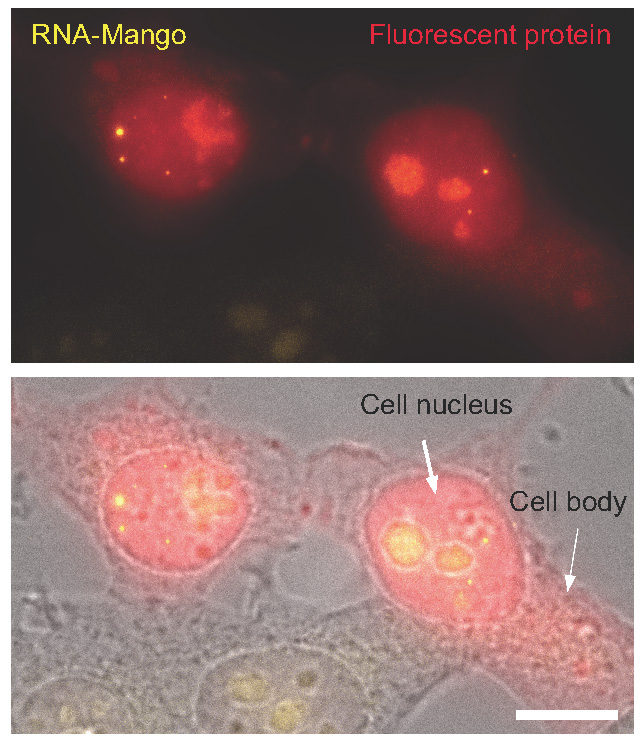
This RNA imaging technique involves incorporation of a synthetic RNA sequence to the RNA of interest. The synthetic RNA sequences can be evolved to selectively and efficiently bind a fluorogenic dye, which, once bound switches on fluorescence. It has been found that the binding of this dye enables a 1000 fold, RNA specific increase in a yellow fluorescent signal. The RNA tags were termed RNA-Mango due to the yellowish-orange colour.
The Canadian team were instrumental in developing the flurogenic dye binding Mango RNA libraries. The French team used advanced microfluidic screening to isolate the new Mango sequences with enhanced imaging properties, making them brighter and ten-times longer lived than previously reported and thus more applicable for imaging cellular RNAs. Adam Cawte, a graduate student at LMS under Professor Rueda’s supervision, demonstrated the applicability of the technique by directly imaging several cellular RNAs.
The LMS team imaged three small RNA molecules that participate in different cellular processes: 5S RNA, involved in translation; U6 RNA, required for splicing; and a scaRNA, which acts in RNA editing. The team provided evidence that the tagged RNAs are localised to their native environment in living cells. The sensitivity is so high that it enables tracking RNA molecules in cellular Foci, meaning that it is possible to monitor where the RNAs go to carry out their function, how long it takes them to get there, and how long they stay there. Furthermore, they showed that the new aptamers, molecules of RNA with dye binding capability, could be used to count the number of molecules in the foci.
The new RNA Mango aptamers can be used to image RNAs at much higher resolution, sensitivity, contrast and longer dye lifetime than previously reported aptamers. This suggests the potential to be widely applied to studying cellular RNAs in ways that have previously not been possible.
This is a very promising imaging technique and the team at LMS hopes that it will become broadly utilised by many other institutes and research laboratories.
The full paper can be read here.