Research at the LMS
The LMS has 29 research groups, each focusing on a different area of medical science.
Our research priorities
We are focused on three main areas:
Our translational target areas
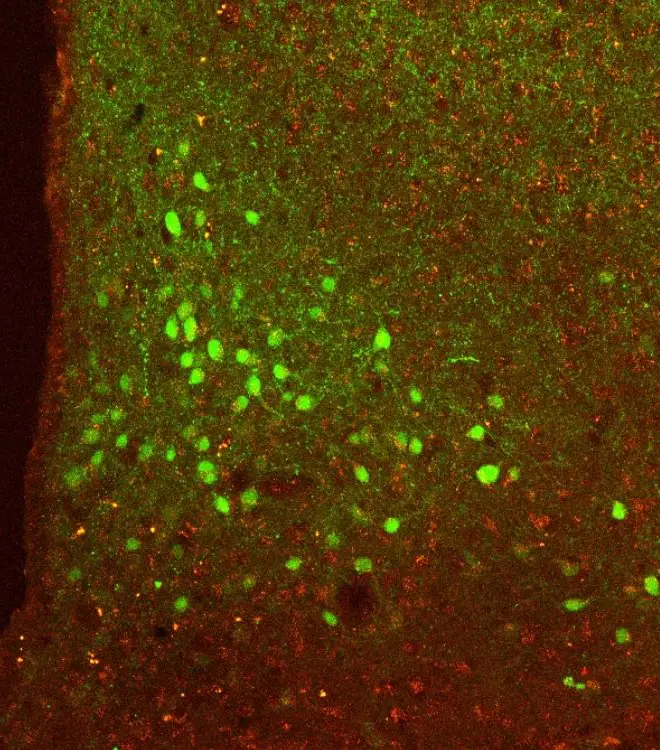
Our research teams work to understand the mechanisms underlying multimorbidity and ageing. We do this to find new diagnostic tests and treatments. Specific health conditions we are working on include…
Fatty liver disease
Heart failure
Chronic inflammation
Cancer
Cardiovascular disease
Dementia
Disorders of early development
Obesity
Diabetes
Polycystic ovary syndrome
Rare inborn complex disease
Challenge-led strategy

Our strategy is focused on team-powered discovery science. Our researchers work flexibly across many disciplines including with external collaborators.
Meet our research groups
Our facilities
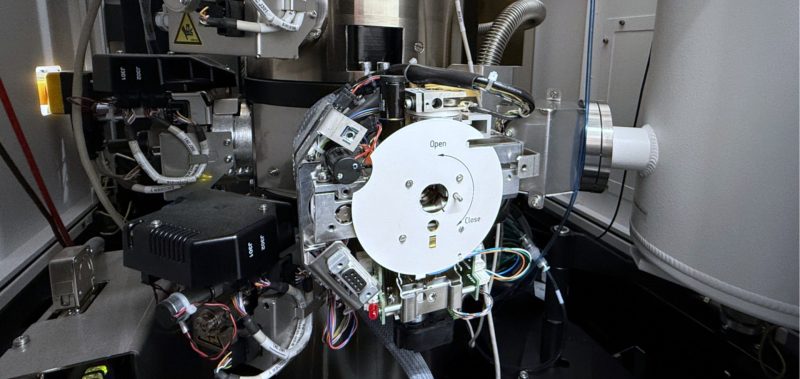
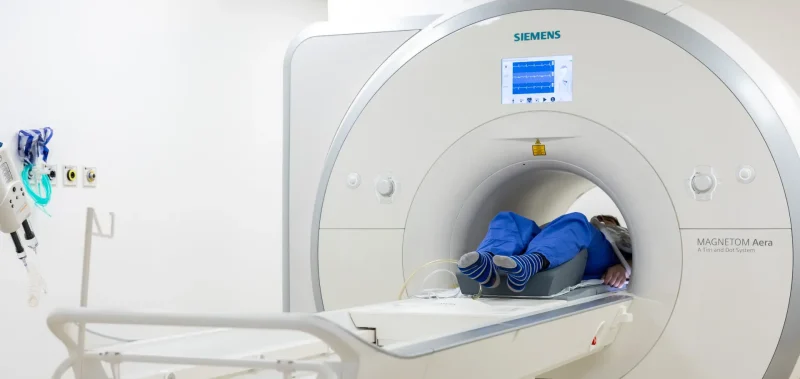
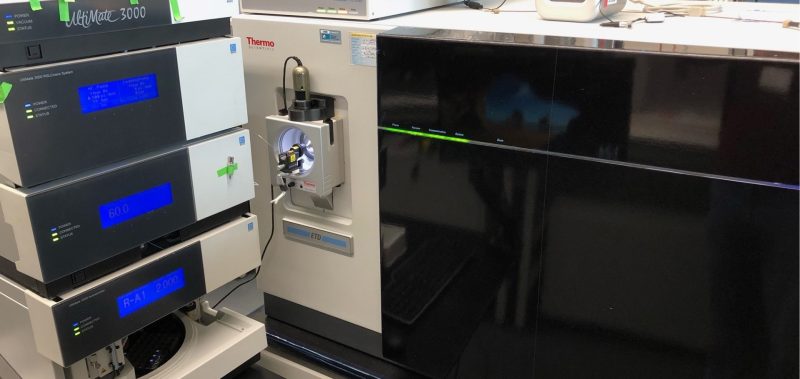
We’re proud of our state-of-the-art facilities that enable us to do groundbreaking research.
Our partners



Latest research news
Study reveals the hidden benefits of weight loss on fat tissue
Published 9 July 2025
3 Minutes reading time