Our laboratory is dedicated to unraveling the intricate mechanisms by which cells organize and manage their extensive DNA molecules. Our primary focus is on understanding the roles of Structural Maintenance of Chromosomes (SMC) complexes and topoisomerases. These essential factors are crucial for the regulation of DNA and chromosomes, impacting a wide range of DNA-centric processes such as transcription, replication, repair, and chromosome segregation. Our ultimate goal is to translate this fundamental knowledge into the development of novel therapeutic strategies targeting these components in various diseases, including cancer and viral infections.
“Can we target certain proteins to treat infections?”
In our laboratory we study SMC complexes and topoisomerases. These are essential tools in our cells that help manage and organize our DNA, ensuring it functions properly. SMC complexes keep our DNA neatly packaged and help during cell division and repair, while topoisomerases act like specialized scissors and glue, fixing the twists and tangles in our DNA. SMC complexes are also involved in defending our cells against some viruses. Understanding how these proteins work is crucial for developing new treatments for diseases like cancer, genetic disorders, and infections. For example, by targeting these factors, we can stop cancer cells from dividing uncontrollably or enhance DNA repair in genetic diseases.
In our lab, we use complementary disciplines like biochemistry, biophysics, and molecular biology to fully understand how SMC complexes and topoisomerases work in our cells. With this knowledge, we are also developing new drugs that will target these proteins, either by blocking or enhancing their functions. These drugs have the potential to be used as therapies to treat cancer and chronic viral infections like hepatitis B.
By understanding the molecular function of SMC complexes and topoisomerase, we can develop drugs that better manage DNA processes. This can lead to more effective treatments for cancer, genetic disorders, and infections, making it a critical area of research in developing new therapies.
Understanding the structure and function of DNA in cells is necessary for deciphering the molecular mechanisms underlying life’s fundamental processes, from gene expression and cellular differentiation to disease development and therapeutic intervention. Structural Maintenance of Chromosomes (SMC) complexes and Topoisomerases intricately regulate the architecture and activity of the genome, exerting profound influences on cellular processes crucial for life and development.
SMC complexes, including condensin, cohesin, and Smc5/6, play indispensable roles in maintaining chromosome structure, ensuring faithful chromosome segregation during cell division, and orchestrating DNA repair processes. Complementing the function of SMC complexes, Topoisomerases are essential enzymes that modulate DNA topology, enabling crucial processes such as DNA replication, transcription, and chromosome condensation. Understanding the intricate mechanisms and regulatory networks governing SMC complexes and Topoisomerases not only sheds light on fundamental aspects of cell biology but also holds promising implications for addressing numerous human health challenges. Our research group employs biochemical, biophysical, single-molecule and in vivo techniques to unravel the complexities of these key molecular players, aiming to uncover novel insights into genome maintenance and contribute to the development of therapeutic strategies.
Our research questions include:
How do SMC complexes interact with DNA and chromatin? We have meticulously purified all three SMC complexes from human cells and endeavour to elucidate their behaviour on different types of DNA (single-stranded and double-stranded) and nucleosome arrays. Through a fusion of single-molecule approaches and biochemistry, we seek to identify the activities and regulatory mechanisms of these molecular machines.
How are the activities of SMC complexes and Topoisomerases coordinated?
Our investigation focuses on deciphering how topoisomerases modulate DNA supercoiling and resolve DNA entanglements in the presence of SMC complexes. Leveraging optical tweezers, we measure changes in DNA topology and mechanical properties. Furthermore, we explore the functional interplay between SMC complexes and topoisomerases during pivotal processes such as DNA replication and repair, utilizing cellular models to validate our findings.
Can we develop novel therapeutics to exploit the translational potential of SMC complexes? Individuals afflicted with SMC5/6-deficient function suffer from a spectrum of genome instability syndromes, manifesting in diverse clinical phenotypes including cardiovascular, hepatic, and pulmonary disorders. Moreover, human SMC5/6 serves as a pivotal host restriction factor against numerous viruses (HBV, EBV, HIV, etc.), many of which harbor oncogenic potential. Exploiting this vulnerability, we are actively engaged in the development of inhibitors to disrupt viral-SMC interactions and modulate SMC function, thereby laying the groundwork for pioneering therapeutic interventions.
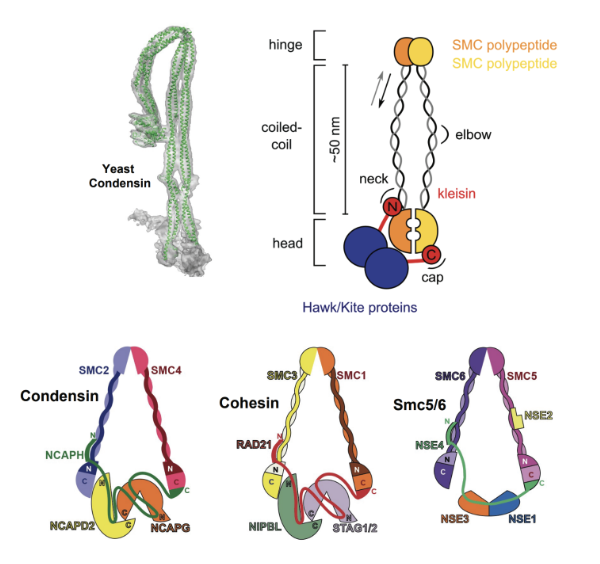
Fig. 1. SMC (Structural Maintenance of Chromosomes) complexes are essential protein assemblies involved in chromosome organization and dynamics. These complexes consist of SMC proteins forming dimers through their hinge domains, which creates a characteristic V-shaped structure. Each SMC protein contains N-terminal ATPase heads (heads) responsible for energy-dependent activities, such as ATP hydrolysis, bridged by a kleisin subunit. Non-SMC subunits (Hawks/Kites) are also present, contributing regulatory and structural roles. SMC complexes interact with DNA via specific DNA-binding regions within the SMC proteins or associated subunits, facilitating their roles in chromosome maintenance and dynamics during cellular processes such as DNA replication, repair, and segregation. Eukaryotic cells contain three distinct classes of SMC complexes, Condensin, Cohesin and Smc5/6. 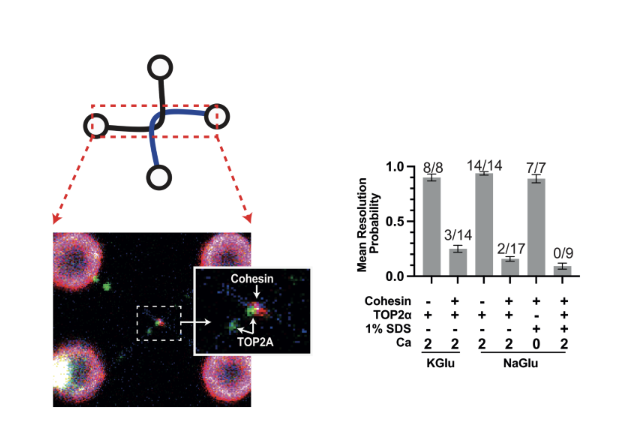
Fig. 2. Human cohesin complex prevents efficient decatenation of DNA entanglements by human Topoisomerase 2 Alpha (TOP2α). Cohesin (red) and TOP2A (green) localise to DNA entanglements (catenations) generated using optical tweezers (left). DNA resolution after incubation with cohesin and TOP2α is checked by removing proteins with 1%SDS (right graph). Mean proportion of DNA resolution in experiments with cohesin and TOP2α, demonstrates that cohesion presence at the DNA catenation inhibits TOP2α-dependent resolution (right).
Our research aims at bridging fundamental genome biology and translational medicine. We aim to understand how proteins involved in genome organisation interact with DNA to preserve genome integrity.
We focus on the Structural Maintenance of Chromosomes (SMC) complexes (cohesin, condensin, and Smc5/6) and on topoisomerases. We employ a combination of biochemical, biophysical, single-molecule, and cellular approaches, to uncover how these molecular machines maintain chromosomal architecture and repair DNA damage.
Translationally, our work is relevant for developmental disorders and cancer, where these complexes malfunction. We are also investigating DNA sensors, such as cGAS, which detect cytoplasmic DNA and trigger innate immunity responses, and consequently this work is relevant to autoimmunity, inflammation, and ageing. Through highthroughput biochemical screens, we aim to identify inhibitors of cGAS and viral modulators of SMC complexes, opening routes to novel antiviral and anti-inflammatory therapies rooted in a deep mechanistic understanding of genome–protein modulators.


Gutierrez-Escribano P, Hormeño S, Madariaga-Marcos J, Solé-Soler R, O’Reilly F, Morris K, Aicart-Ramos C, Aramayo R, Montoya A, Kramer H, Rappsilber J, Torres-Rosell J, Moreno-Herrero F, Aragon L. (2020). Purified Smc5/6 Complex Exhibits DNA Substrate Recognition and Compaction. Molecular Cell 80 (6), 1039-1054.
Lee B-G, Merkel F, Allegretti M, Hassler M, Cawood C, Lecomte L, O’Reilly F, Sinn L, Gutierrez-Escribano P, Kschonsak M, Bravo S, Nakane T, Rappsilber J, Aragon L*, Beck M*, Löwe J*, Haering C.* (2020). Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nature Structural and Molecular Biology 27, 743-751.
Bardell-Cox O, White AJP, Aragon L*, Fuchter MJ. * (2019). Synthetic studies on the reverse antibiotic natural products, the nybomycins. Med Chem Commun 10 (8), 1438-1444.
Gutierrez-Escribano P, Newton M, Llauro A, Huber J, Tanasie L, Davy J, Aly I, Aramayo R, Montoya A, Kramer H, Stigler J, Rueda D, Aragón L. (2019). A conserved ATP and Scc2/4-dependent activity for cohesin in tethering DNA molecules. Science Advances 5 (11), eaay6804.
Garcia-Luis J, Lazar-Stefanita L, Gutierrez-Escribano P, Thierry A, Garcia A, Sanchez M, Jarmuz A, Montoya A, Dore M, Kramer H, Karimi M, Antequera F, Koszul R, Aragón L. (2019). FACT mediates cohesin function on chromatin. Nature Structural and Molecular Biology 26, 970-979
Sen N, Leonard J, Torres R, Garcia-Luis J, Palou-Marin G, Aragón L. (2016). Physical Proximity of Sister Chromatids Promotes Top2-Dependent Intertwining, Molecular Cell 64(1), 134-147.