The genomic variation and disease team at the LMS started in 2024. Led by Lila Allou, it aims to understand the molecular consequences of SVs affecting noncoding regions in development and disease, thereby improving rare disease diagnosis and identifying potential therapeutic targets in cancer.
“How do changes in the structure of chromosomes affect health and development?”
In the nucleus of each cell, a DNA molecule forms thread-like structures called chromosomes. Chromosomes are thus a collection of genes carrying genetic information.
Chromosome mutations, which can modify the number or structure of chromosomes in a cell, are a complex phenomenon. While a gene mutation affects only a single gene on a chromosome, a chromosomal mutation can disrupt the function of more than one gene and/or impact the noncoding DNA sequences located between genes. These noncoding regions have crucial roles in regulating the amount of proteins that are produced. Chromosome mutations can thus lead to issues with the growth, development, and function of body systems.
Our team is researching exactly how changes that affect chromosome structure can cause rare genetic diseases. We are also researching potential therapeutic targets in cancer derived from changes that affect chromosome structure.
By deepening our understanding of the consequences of changes that affect chromosome structure, we have the potential to make a significant impact. This knowledge could lead to identifying the genetic cause of rare diseases, which is of huge benefits to both patients and their families. This knowledge could also lead to the development of innovative therapeutic strategies for cancer patients.
“Structural variation impacting noncoding DNA sequences is a critically understudied class of mutation, although it is likely to contribute significantly to unsolved genetic disease.”
Structural variants (SVs) are changes in the genome structure with a minimum size of 50 bp. They can include inversions, balanced translocations, or genomic imbalances (insertions, deletions, and duplications). Interpreting the impact of SVs, particularly those impacting noncoding DNA sequences, on human health and disease is challenging. Nonetheless, this is of particular importance to diagnose at least a subset of the 300 million undiagnosed patients with a rare disease and to develop novel therapeutic strategies for cancer patients.
Our research aims to unravel novel SV-associated molecular consequences with the purpose of contributing valuable insights for rare disease diagnosis and cancer treatment. We plan to:
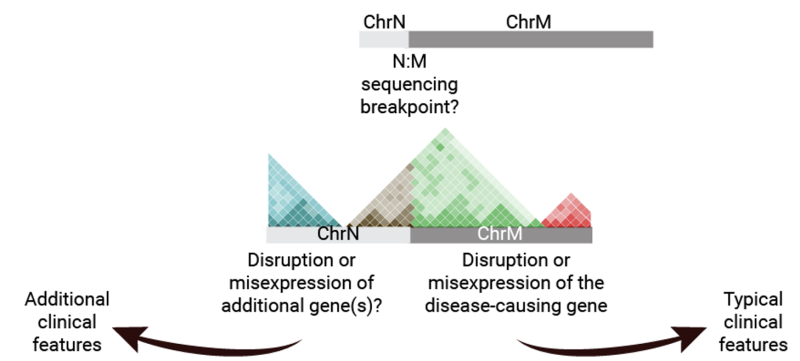
The knowledge gained from our research could lead to the diagnosis of a substantial portion of the 300 million undiagnosed patients with rare diseases and the development of innovative therapeutic strategies for cancer patients.
In particular, our research aims to deepen our understanding of the functional consequences of genetic structural variations (SVs), the development of new tools for the clinical interpretation of SV effects, and the characterisation of potential SV-derived therapeutic targets in cancer.
Our lab focuses on investigating SVs impacting noncoding DNA sequences and uses a combination of mouse and human models, genome-editing technologies, single-cell sequencing, and proteomics.

Alessa R. Ringel*, Andreas Magg*, Natalia Benetti, Robert Schöpflin, Mira Kühnlein, Asita Carola Stiege, Ute Fischer, Lars Wittler, Stephan Lorenz, Stefan Mundlos#, Lila Allou#. Temporal misexpression of En1 during limb development causes distinct phenotypes. bioRxiv. 2024 Sep. (* joint first authors, # joint corresponding authors)
Allou L#, Mundlos S#. Disruption of regulatory domains and novel transcripts as disease-causing mechanisms. Bioessays. 2023 Oct;45(10):e2300010. (# joint corresponding authors)
Allou L, Balzano S, Magg A, Quinodoz M, Royer-Bertrand B, Schöpflin R, Chan WL, Speck-Martins CE, Carvalho DR, Farage L, Lourenço CM, Albuquerque R, Rajagopal S, Nampoothiri S, Campos-Xavier B, Chiesa C, Niel-Bütschi F, Wittler L, Timmermann B, Spielmann M, Robson MI, Ringel A, Heinrich V, Cova G, Andrey G, Prada-Medina CA, Pescini-Gobert R, Unger S, Bonafé L, Grote P, Rivolta C, Mundlos S, Superti-Furga A. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature. 2021 Apr;592(7852):93-98. (F1000 Prime recommended paper, PNAS Journal Club)
Allou L, Julia S, Amsallem D, El Chehadeh S, Lambert L, Thevenon J, Duffourd Y, Saunier A, Bouquet P, Pere S, Moustaïne A, Ruaud L, Roth V, Jonveaux P, Philippe C. Rett-like phenotypes: expanding the genetic heterogeneity to the KCNA2 gene and first familial case of CDKL5-related disease. Clin Genet. 2017 Mar;91(3):431-440.
Allou L*, Lambert L*, Amsallem D, Bieth E, Edery P, Destrée A, Rivier F, Amor D, Thompson E, Nicholl J, Harbord M, Nemos C, Saunier A, Moustaïne A, Vigouroux A, Jonveaux P, Philippe C. 14q12 and severe Rett-like phenotypes: new clinical insights and physical mapping of FOXG1-regulatory elements. Eur J Hum Genet. 2012 Dec;20(12):1216-23. (* joint first authors)