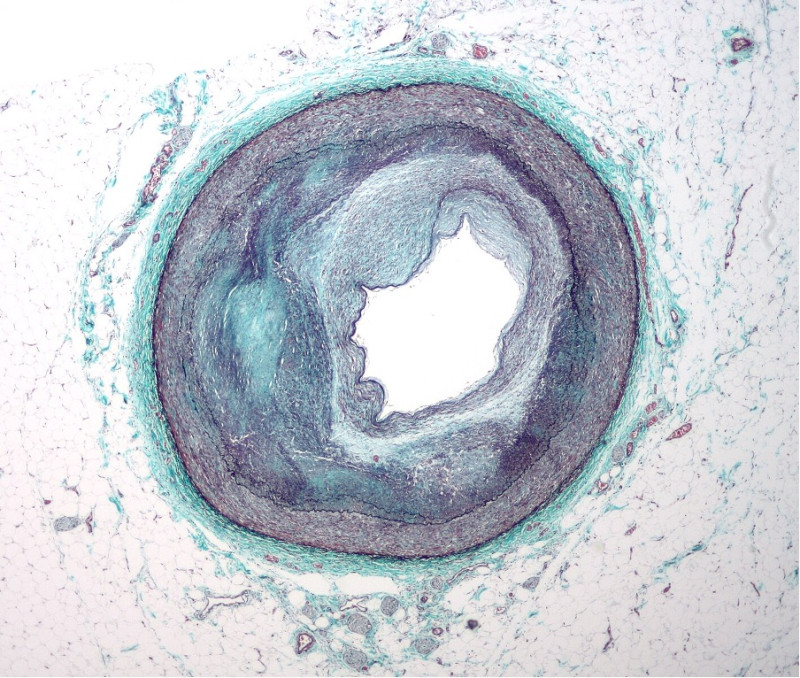
Vascular smooth muscle cells (VSMCs) are found in the walls of blood vessels and play a critical role in maintaining correct blood pressure and blood flow. If the blood vessels become injured or inflamed, some of these cells lose their muscle cell features, migrate, and start dividing to promote healing. However, if this process is uncontrolled, it can result in atherosclerosis. Atherosclerosis is the buildup of plaques in blood vessels which causes them to stiffen, and is the main cause of cardiovascular diseases and strokes. This makes it the underlying cause of death in about 50% of populations in the Western world. Dividing VSMCs are the primary component of the atherosclerotic plaques building up in blood vessels, which is why we critically need to understand what factors push these cells to transition from their contractile (muscle-like) state to the actively dividing state.
In a study published in Nature Cardiovascular Research, a collaboration of scientists from Mikhail Spivakov’s lab (MRC LMS) and Helle Jørgensen’s lab (University of Cambridge) have defined the key genes that control the switching of VSMCs to their dividing state. To do this, they combined extensive experimental and computational techniques. These findings may help better understand how to reduce cardiovascular disease risk and provide avenues towards finding preventative measures.
Prior work by Spivakov’s and Jørgensen’s labs identified a population of VSMCs that expressed a gene called Sca1 which had a higher capacity to proliferate after vascular injury. Subsequently, the researchers found that rather than being a distinct cell type, these cells were in transition between their contractile to the actively dividing state. In their new paper, the teams have captured the full complexity of VSMC inflammatory activation, using powerful single-cell transcriptomics and epigenetic profiling techniques. “In this study, we focus on the transitioning VSMCs that we have characterised previously and use exciting computational methodologies to really dig into the mechanisms that control this process” – said Mikhail Spivakov, the group leader at the LMS.

Single-cell transcriptomics and epigenetic profiling involve researchers looking at each individual cell in a group, to extract information on how it functions. Transcriptomics informs scientists on the RNA content of the cell, while epigenetic profiling is a tool to examine the regulation of DNA within each cell. The organisation of DNA tells the cell which instructions to follow so it can function correctly.
Based on these data, researchers then built gene regulatory networks. A gene regulatory network represents roadmaps for how cells make decisions, by plotting out which genes are turned on (or off) according to the signals they receive. The researchers could then model the behaviour of the cells upon perturbing different components of the network computationally, and then examine the impact of these perturbations. This computational analysis has enabled the teams to pinpoint the key factors that guide VSMC response, providing avenues into experimental validation. “We identified gene regulatory networks driving transcriptional changes towards VSMC activation, and through computational analyses and simulation of these networks, we could prioritize candidate factors for further investigation” – said Sebnem Oc, a postdoc in the Jørgensen lab who is co-affiliated with Spivakov’s team at the LMS.

Combining network analysis with targeted experiments, the researchers identified several key factors and pathways regulating VSMC disease response. These included the transcription factor Runx1, which is involved in activating the expression of other genes and is known to be related to VSMC function. TIMP1 was also noted to be important in VSMC switching, which is a protein involved in regulating signals for tissue remodelling. The experiments to validate these factors were done in a mouse vascular injury model. Finally, the researchers then used patient biopsies of atherosclerotic plaques to show that the same factors likely guide VSMC inflammatory response in humans.
Atherosclerosis is estimated to affect almost 7% of adults in the UK and underpins heart attacks and stroke. Insights from the gene regulatory network analysis open promising doors to understanding the mechanisms by which VSMC proliferation links to eventual atherosclerotic plaque build-up and can inform future strategies for preventing and treating this common and complex disease. “The hope is that our findings will help combat the development of cardiovascular disease, and contribute to designing new strategies for treatment” – said Helle Jørgensen, the study’s senior author.

This work was funded by the British Heart Foundation, the Cambridge and Leicester NIHR Biomedical Research Centres, the Deutsche Forschungsgemeinschaft, the Medical Research Council of the UK, the Chan Zuckerberg Initiative, Singapore’s National Medical Research Council, and National University of Singapore and National University Health System and has now been published in Nature Cardiovascular Research.
By Srishti Arya



