By Helen Figueira
May 31, 2016
Time to read: 5 minutes
By Susan Watts
Scientists are developing ever more detailed models to explain how DNA is stored inside our cells, and why it adopts the shapes it does.
In a review of the latest research in the field Matthias Merkenschlager, of the CSC’s Lymphocyte Development group, and joint author Elphege Nora, of the Gladstone Institute of Cardiovascular Disease in San Francisco, examine the layers of order within our genome, beyond segmentation into chromosomes. In their paper, published online in the Annual Review of Genomics and Human Genetics, they discuss remaining questions and how future research might tackle these.
At the highest level, our genome is segmented into chromosomes. But it doesn’t stop there. There’s a further level of segmentation, known to be crucial for the healthy functioning of the cell. This segmentation occurs through folding of the DNA. Each resulting domain becomes a functional unit, and these domains are important for maintaining key aspects of a healthy cell, as it divides to make copies of itself and expresses the information encoded in its genome’s sequences.
The dynamic process of folding and looping of the chromosomal DNA is exquisitely choreographed, bringing together regions of the DNA that are not normally adjacent. Mistakes in this process can have serious consequences, and have been linked to rare genetic disorders and certain types of cancer, including colorectal cancer and acute myeloid leukaemia.
Segmentation of the genome into domains matters for three key reasons. First, it helps to ensure that the cell copies its own DNA faithfully during DNA replication. Second, it helps to prevent genetic re-arrangements, by physically separating similar stretches of DNA that if they were to combine could cause a problem. And third, it simplifies gene regulation. We have around 20,000 genes and some 100,000 enhancers. If all of those genes and enhancers could pair randomly, there would be a huge number of potential combinations. In this scenario, random enhancers would drive the expression of random genes. The idea is that if only a small number of genes and a small numbers of enhancers are placed in the same domain, they find each other and regulation becomes a simpler task.
“There’s this catchphrase ‘spatial segmentation translates into regulatory segmentation’, which simply means that if regulatory elements are separated in space they wont influence each other,” Merkenschlager explains. In other words, elements like enhancers do not interact randomly across the genome. Instead they preferentially interact with each other if they are in the same domain.
The details of how exactly segmentation by folding happens remain unknown. It’s apparent, however, that two players, called CTCF and cohesin, have important roles. Cohesin is a ring-like protein complex that encircles looping strands of DNA and holds them in place. A second protein, CTCF, helps to ensure that cohesin binds to DNA in the right place.
One of the most interesting and potentially most significant constraints on a mechanism to explain loop formation is the recent observation that the way that loops are formed depends on the orientation of the DNA sequence that binds CTCF. Interacting sequences are preferentially lined up to “look at each other”.
“It’s like reading. It’s a processive event in which it matters where you started and which way you are going.” Merkenschlager says. “That observation was startling, like a throw-back to the 1980s where everything – transcription, replication – they all have a direction. Until now, looping was not thought to have a direction, it was always a fluffy thing and now suddenly looping had a direction, and that’s super interesting.”
One intriguing but still speculative scenario for how CTCF and cohesin work together to accomplish genome folding is called the “loop extrusion model”. To date, however, there’s no accepted mechanism for exactly how this model might work at the molecular level, though people have visualised various possibilities. All give directionality to sequences by extruding a loop through a nozzle or ring, such that the DNA emerges aligned.
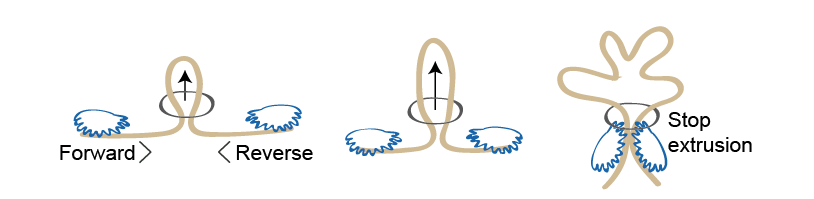
The model appeals to many in the field not only because it fits observation. “Fundamentally it’s useful, because if you can extrude the DNA through a ring it means this DNA is not knotted up, it’s smooth, unperturbed and structurally there’s no problem.”
The authors have published extensively on the nature of domains in the genome, and on the roles of CTCF and cohesin, but not on loop extrusion per se, so are well placed to stand back and review the various proposed interpretations. “We’ve run these past all the data that we have about genome folding, and its significance for gene regulation and replication, and in this review we’re saying which are the sensible assumptions and which are the not so sensible assumptions.”
Read more on this latest review – first posted online on 18 April 2016
Contact
Susan Watts
Head of Communications and Public Engagement
MRC Clinical Sciences Centre
L: 0208 383 8247
M: 07590 250652
E:
Further reading on Cohesin, CTCF and the loop extrusion model