By Sofia Velasquez-Pimentel
March 20, 2023
Time to read: 6 minutes
How repressive chromatin maintain cell identity
By Sofia Velazquez Pimentel
Ground-breaking findings from the MRC London Institute of Medical Sciences (LMS) have revealed that the repressive epigenetic mark “Histone H3K9 trimethylation is essential for maintaining mitotic chromosome structure and ‘DNA bookmarking’ during cell division, ensuring that ‘cell identity’ is accurately passed on from mother to daughter cells. Investigating these nuances on how cells maintain the memory of their ‘identity’ will have critical implications in human development and diseases such as immune disorders and cancer.
Every cell in our body contains the same DNA, yet there are over two hundred different cell types with their own unique structure and function. This uniqueness is guided by different sets of genes, which can be turned ‘on’ or ‘off’ to create a distinctive signature and determine cell identity. These genes are switched on or off through epigenetic changes that affect how easily DNA can be ‘read’ and a gene can be expressed (or functionally active). This essential process underlies the transition of cells into specialized cell types, such as immune, muscle, or skin cells. In their newest publication, LMS researchers provide new insights into the role of repressive machinery that ‘blocks’ DNA from being ‘read’ in maintaining cell identity and carries important implications for understanding diseases such as cancer.
Understanding the mechanisms underlying cell memory is crucial for comprehending the stability and plasticity of cells in normal physiological processes and diseases. For instance, the ability to reverse the memory of cancer cells with a ‘forget mechanism’ can be a promising therapeutic strategy in the future.
In preparation for cell division, DNA is tightly wound into chromosomes and DNA is ‘bookmarked’ so that the cell can ‘remember its identity’. In order to ensure cellular memory and keep cell identity through cell division, cells need first to duplicate their genetic and epigenetic material onto the newly synthetized DNA strand during replication, then maintain this information during mitosis phase of the cell cycle until appearance of two identical daughter cells.
‘Conveying epigenetic memory and cellular identity though cell division is a fundamental question that is just beginning to become understood and I hope that this study and the methodologies that we have developed with the incredible work of Bhavik Patel and Holger Kramer will contribute to better understand and address the epigenetic memory questions.’
– Dr Dounia Djeghloul, first and co-corresponding author andresearcher in the LMS Epigenetic Memory group.
During mitosis, chromosomes undergo extensive structural changes and become highly condensed. For a long time, it was thought that a lot of this information copied during the DNA replication were lost during mitosis. This made it extremely challenging to understand how cells were able to sustain their identity as they divide.
To tackle this problem, Pr. Amanda Fisher -leader of the LMS Epigenetic Memory group – previously developed a technology combining flow cytometry and high throughput proteomics which allowed researchers to isolate chromosomes from cells with minimal damage so that they were near identical to how they were in the cell. This cutting-edge technique allowed the team to identify the factors and information contained on native mitotic chromosomes and test their functional importance (Djeghloul et al 2020).
In the team’s latest publication, Djeghloul et al., 2023 addressed the link between chromatin state and mitotic bookmarking. They found that a repressive epigenetic mark histone H3K9 methylation, which blocks DNA from being ‘read’ is essential for sustaining retention of ‘bookmarking factors’ during cell division (particularly bookmarking by the transcription factor Esrrb in mouse embryonic stem cells).
Researchers were surprised to discover that the removal of H3K9 methylation led to an over compaction of mitotic chromosomes. This suggests that this repressive epigenetic mark also plays a pivotal role in maintaining mitotic chromosome structure.
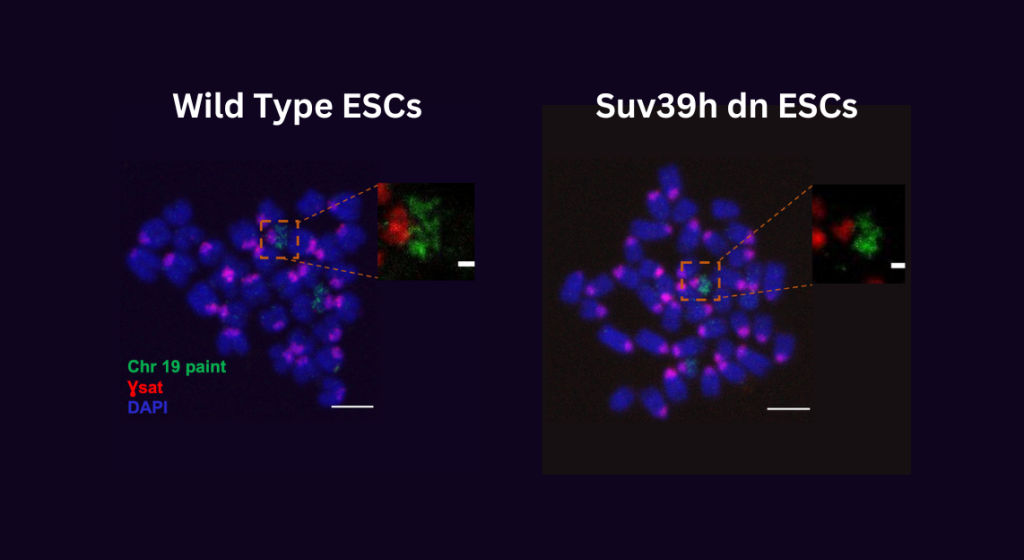
Researchers were shocked to find that mutant cells lacking H3k9 trimethylation (right image) have extremely small and compact chromosomes arms (green) and centromeres (red)compared to wild-type cells (left image).
‘This study addresses a new link between chromatin state and mitotic bookmaking. Counter-intuitively repressive chromatin mark H3K9 methylation prevent over compaction of mitotic chromosomes and help retention of bookmarking factors on mitotic chromosomes during cell division’ explained Dr Dounia Djeghloul, first and co-corresponding author ‘conveying epigenetic memory and cellular identity though cell division is a fundamental question that is just beginning to become understood and I hope that this study and the methodologies that we have developed with the incredible work of Bhavik Patel and Holger Kramer will contribute to better understand and address the epigenetic memory questions’.
This study highlights an unexpected role for repressive chromatin in maintaining epigenetic memory and cellular identity through cell division and carries important implications for understanding human development and diseases such as immune disorders and cancer.
‘This research has been a fantastic collaboration within the LMS. We would not have this paper without LMS’ people and facilities; special mention is owed to Bhavik Patel from the Flow Cytometry facility, who isolated the chromosomes, and Holger Kramer from the Proteomics facility, who analysed the chromosomes by mass spectrometry.’
– Dr Andrew Dimond, second-author and and researcher in the LMS Epigenetic Memory Group.
‘This exciting study by Mandy, Dounia and the team helps to answer fundamental questions as to how daughter cells remember their identity which has great significance in the understanding of autoimmune diseases and cancer. It was an absolute pleasure to have been given the opportunity to contribute to this project’
-Dr Bhavik Patel, co-author and Technical Director of the LMS Flow Cytometry Facility.
‘This fantastic piece of work shows that different flavours of chromatin influence the retention of transcription factors at mitotic chromosomes – something that was not previously known’
– Professor Dame Amanda Fisher, lead author and co-corresponding author and leader of the LMS Epigenetic Memory Group.
This research was published in Nature Structural & Molecular Biology and was produced by the MRC LMS Epigenetic Memory group, with the highly valuable help from MRC LMS Biological Mass Spectrometry and Proteomics, MRC LMS Flow Cytometry, MRC LMS Microscopy, and MRC LMS Bioinformatics, MRC LMS Lymphocyte Development and in collaboration with the Max-Planck Institute of Immunobiology and Epigenetics.
This research was funded by Medical Research Council UK and The Wellcome Trust Institutional Strategic Support Fund Springboard award.
Read the Nature paper HERE