
Amanda Fisher
Group Members
Lab Admin Contact
Funding


“Understanding how epigenetic information is transmitted and changed during normal development and in disease.”
Our team examines how cellular identity is established, progressively changed during differentiation and then transmitted through cell division. We also study how environment exposures including those encountered in utero, can impact the developing epigenome and thereby influence the health of offspring during their lifetime and across generations.
Understanding how epigenetic information is transmitted from mother to daughter cells is critical for understanding cellular inheritance. As cells enter mitosis, chromosomes condense ahead of a breakdown of the nuclear envelope. Some DNA-associated factors are displaced from mitotic chromosomes while others remain bound. It has been proposed that these chromosome-bound factors are critical for ‘bookmarking the genome’ to ensure that cell identity is correctly reinstated and passed to daughter cells following cell division.
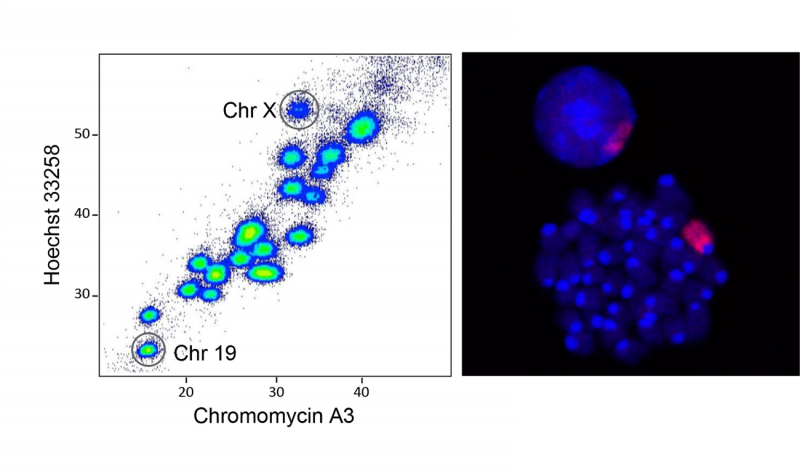
Flow sorting of mouse chromosomes 19 and X (left) and visualizing the X chromosome in mitotic cells (red, right).
We are developing convergent approaches to systematically identify factors such as proteins and RNAs that remain bound to isolated mitotic chromosomes, as well as probing their functional relevance. Current studies focus on comparing mitotic bookmarking in different stem cell types and their differentiated progeny, understanding the dependency of bookmarking on underlying chromatin features, and determining the mitotic features of active and inactive X chromosomes that are important in maintaining their states through cell division. Long-term this information will be used to design novel epigenetic strategies aimed at altering cellular identity or changing gene expression for therapeutic benefit.
A second theme of our research examines how chromatin-modifying drugs or exposures impact the developing epigenome, using bespoke bioluminescent imaging designed to report the allelic expression of genes in living systems and in pre-clinical models. These studies reveal how exposures can alter epigenetic responses (using reporters for imprinted or X-linked genes), developmental ontogeny (using Utrn and Dmd) reporters and signalling (for example using Cyp1a1 as an indicator for environmental pollutants such as dioxin).
Long-term this information is important to better understand how genetic, lifestyle and the environmental factors shape our health, and to design therapeutic interventions that can harness the re-expression of epigenetically-silent genes to correct or ameliorate disease.
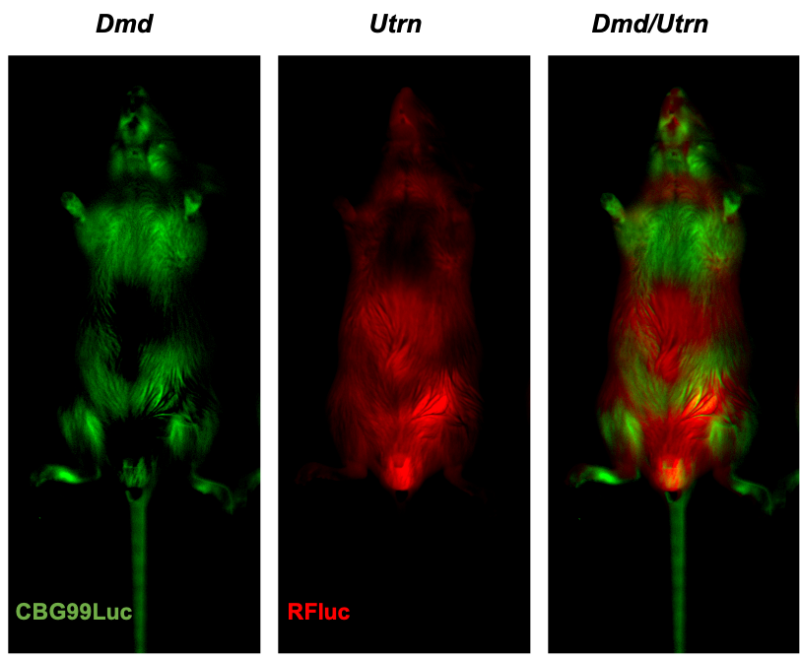
Spectral unmixing of bioluminescent signals in a novel dual reporter line (Dmd-CBG99Luc/Utrn-Rfluc) reveals gene expression in vivo.
Selected Publications
Van de Pette M, Dimond A, Galvao A, Millership SJ, To W, Prodani C, McNamara G, Bruno L, Sardini A, Webster Z, McGinty J, French P, Uren AG, Castillo-Fernandez Juan, Watkinson William, Ferguson-Smith AC, Merkenschlager M, John RM, Kelsey G & Fisher AG. (2022) Epigenetic changes induced by in utero dietary challenge result in phenotypic variability in successive generations of mice, Nature Communications, 2022 May 13, 2464. doi: 10.1038/s41467-022-30022-2.
Djeghloul D, Dimond A, Kramer H, Brown K, Patel B, Wang Y-F, Futschik ME, Whilding C, Montoya A, Veland N, Cheriyamkunnel S, Montavon T, Jenuwein T, Merkenschlager M, Fisher AG. (2022) Loss of H3K9 tri-methylation alters chromosome compaction and transcription factor retention during mitosis bioRxiv 2022.02.01.478684; doi: https://doi.org/10.1101/2022.02.01.478684
Weiss FD, Calderon L, Wang Y-F, Georgieva R, Guo Y, Cvetesic N, Kaur M, Dharmalingam G, Krantz ID, Lenhard B, Fisher A, Merkenschlager M (2021). Neuronal genes deregulated in Cornelia de Lange Syndrome respond to removal and reexpression of cohesin, Nature Communications, Vol: 12, ISSN: 2041-1723
Karimi MM, Guo Y, Cui X, Pallikonda HA, Horkova V, Dore MH, Wang Y-F, Ruiz Gil S, Rodriguez-Eteban G, Robles Rebollo I, Bruno L, Georgieva R, Patel B, Elliott J, Dauphars D, Krangel MS, Lenhard B, Heyn H, Fisher AG, Stepanek O, Merkenschlager M (2021). The order and logic of CD4 versus CD8 lineage choice and differentiation in mouse thymus, Nature Communications, Vol: 12, ISSN: 2041-1723
Djeghloul D, Patel B, Kramer H, Dimond A, Whilding C, Brown K, Kohler A, Feytout A, Veland N, Elliott J, Bharat TAM, Tarafder AK, Löwe J, Ng BL, Guo Y, Guy J, Huseyin MK, Klose RJ, Merkenschlager M, Fisher AG. (2020). Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction. Nature Communications 11, 4118 https://doi.org/10.1038/s41467-020-17823-z
Dimond A, Van de Pette M, Fisher AG. (2020). Illuminating Epigenetics and Inheritance in the Immune System with Bioluminescence. Trends Immunol. 41 (11), 994-1005. https://doi.org/10.1016/j.it.2020.09.001
Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Irene Robles-Rebollo, Xiao X, Wang Y-F, Barozzi I, Djeghloul D, Amano MT, Niskanen H, Petretto E, Dowell RD, Tachibana K, Kaikkonen MU, Nasmyth KA, Lenhard B, Natoli G, Fisher AG, Merkenschlager M. (2018). Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nature Immunology 19, 932–941. https://doi.org/10.1038/s41590-018-0184-1
Cantone I, Dharmalingam G, Chan Y-W, Kohler A-C, Lenhard B, Merkenschlager M, Fisher AG. (2017). Allele-specific analysis of cell fusion-mediated pluripotent reprograming reveals distinct and predictive susceptibilities of human X-linked genes to reactivation. Genome Biology 18, 1474-760.
https://doi.org/10.1186/s13059-016-1136-4
Van de Pette M, Abbas A, Feytout A, McNamara G, Bruno L, To WK, Dimond A, Sardini A, Webster Z, McGinty J, Paul EJ, Ungless MA, French PMW, Withers DJ, Uren A, Ferguson-Smith AC, Merkenschlager M, John RM, Fisher AG. (2017). Visualizing Changes in Cdkn1c Expression Links Early-Life Adversity to Imprint Mis-regulation in Adults. Cell Rep. 18 (5), 1090-1099. https://doi.org/10.1016/j.celrep.2017.01.010
Cantone I, Bagci H, Dormann D, Dharmalingam G, Nesterova T, Brockdorff N, Rougeulle C, Vallot C, Heard E, Chaligne R, Merkenschlager M, Fisher AG. (2016). Ordered chromatin changes and human X chromosome reactivation by cell fusion-mediated pluripotent reprogramming. Nature Communications 7:12354. https://doi.org/10.1038/ncomms12354
