
Enrique "Fadri" Martinez-Perez
Group Members
Funding

“We investigate mechanisms of genome organisation and inheritance in germ cells to understand the causes of infertility and gamete aneuploidy, a leading cause of miscarriages and birth defects.”
Our group aims to understand the molecular mechanisms that ensure accurate chromosome segregation during meiosis, the specialised cell division programme that produces haploid gametes from diploid germ cells. Defects in this process cause infertility and are a leading cause of miscarriages and birth defects in humans. Using the nematode C. elegans as a model organism and a combination of experimental approaches, our research focuses on how different cohesin complexes and HORMA-domain proteins control the structure and function of meiotic chromosomes. We also investigate the quality-control mechanisms that orchestrate meiotic progression by monitoring meiotic chromosome structure. By focusing on these conserved components of the meiotic programme, our research aims to understand causes of human infertility and aneuploidy.
We are also investigating how cohesin complexes ensure normal organism development by controlling 3D genome organisation and gene expression, as mutations in cohesin are a cause of Cornelia de Lange syndrome.
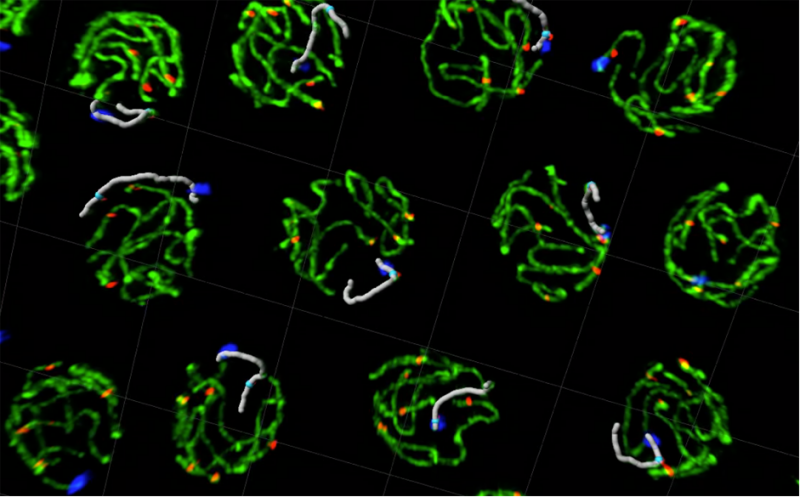
Tracking individual chromosomes in 3D intact pachytene nuclei by super resolution microscopy reveals inter- and intra-nucleus features of chromosome organisation.
Selected Publications
Belan O, Barroso C, Kaczmarczyk A, Anand R, Federico S, O’Reilly N, Newton MD, Maeots E, Enchev RI, Martinez-Perez E, Rueda DS, Boulton SJ. (2021). Single-molecule analysis reveals cooperative stimulation of Rad51 filament nucleation and growth by mediator proteins. Molecular Cell 81 (5), 1058-1073.e7
Castellano-Pozo M, Pacheco S, Sioutas G, Jaso-Tamame AL, Dore MH, Karimi MM, Martinez-Perez E. (2020). Surveillance of cohesin-supported chromosome structure controls meiotic progression. Nature Communications 11 (1), 4345.
Beltran T, Barroso C, Birkle TY, Stevens L, Schwartz HT, Sternberg PW, Fradin H, Gunsalus K, Piano F, Sharma G, Cerrato C, Ahringer J, Martínez-Pérez E, Blaxter M, Sarkies P. (2019). Comparative Epigenomics Reveals that RNA Polymerase II Pausing and Chromatin Domain Organization Control Nematode piRNA Biogenesis. Developmental Cell 48 (6), 793-810.e6.
Link J, Paouneskou D, Velkova M, Daryabeigi A, Laos T, Labella S, Barroso C, Pinol SP, Montoya A, Kramer H, Woglar A, Baudrimont A, Markert SM, Stigloher C, Martinez-Perez E, Dammermann A, Alsheimer M, Zetka M, Jantsch V. (2018). Transient and Partial Nuclear Lamina Disruption Promotes Chromosome Movement in Early Meiotic Prophase, Developmental Cell 45 (2), 212-225.e7.
Ferrandiz N, Barroso C, Telecan O, Shao N, Kim H-M, Testori S, Faull P, Cutillas P, Snijders AP, Colaiacovo MP, Martinez-Perez E. (2018). Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nature Communications 9 (1), 834.
Crawley O, Barroso C, Testori S, Ferrandiz N, Silva N, Castellano-Pozo M, Jaso-Tamame AL, Martinez-Perez E. (2016). Cohesin-interacting protein WAPL-1 regulates meiotic chromosome structure and cohesion by antagonizing specific cohesin complexes. eLife 5: e10851.
