The ability of the peripheral nervous system to repair itself after injury depends on the reprogramming of specialised nerve cells, called Schwann cells, to a more progenitor-like state in order to boost the process of regeneration. A new study published in the latest issue of Neuron now sheds light on the molecular mechanisms that drive this process <https://www.ncbi.nlm.nih.gov/pubmed/28957681>.
The paper is the result of experiments at the MRC Laboratory of Medical Sciences (LMS) where researchers isolated Schwann cells from intact and severed mouse nerves to obtain a full transcriptome of the cells in each state.
Schwann cells are the glia of the peripheral nervous system that myelinate or wrap around axons to accelerate conduction of nerve impulses from brain to limb.
When a nerve is cut, the two cut stumps retract leaving a gap in the tissue, but the damaged axons are able to regrow across the gap due to the ability of the Schwann cells to revert to a stem-like state – in a process called ‘dedifferentiation’. Dedifferentiated Schwann cells divide and migrate to form multicellular cords that bridge the gap to provide paths along which the axons can regrow.
Dr Simona Parrinello, who leads the Cell Interactions and Cancer research group at LMS, said it had been a technical ‘challenge’ involving ‘a lot of trial and error’ to purify sufficient numbers of Schwann cells for molecular analysis of the transcriptional changes and signalling pathways involved in this reprogramming.
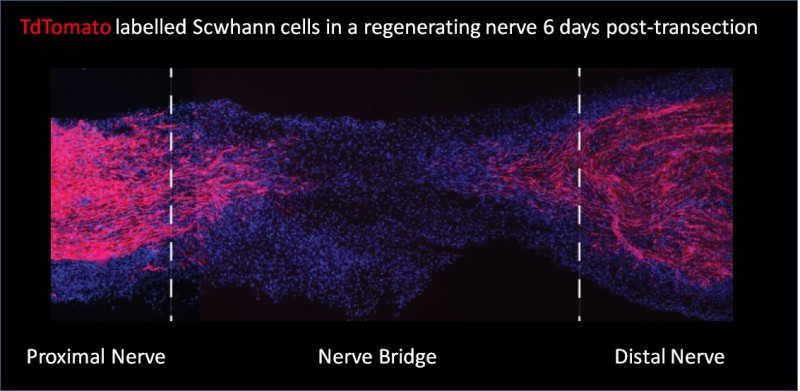
Her team generated a mouse model that selectively labelled Schwann cells with red fluorescent protein, and then used a combination of fluorescence-activated cell sorting and single cell RNA-amplification protocols to carry out transcriptional profiling of in vivo Schwann cells from intact nerves and from the bridge and distal stumps of transected nerves. https://www.youtube.com/watch?v=oJGwVdPArJI
‘Having this transcriptome of the Schwann cells is important because if you can understand the natural mechanisms that drive this remarkable regenerative process in rodents, then you may be able to apply this knowledge to improving nerve repair in humans,’ Parrinello said.
‘Cutting a finger, being in a car accident, these injuries happen to people all the time, and are a big socio-economic burden. Usually the cut nerve stumps are reattached surgically or a nerve conduit is introduced as an artificial means of guiding axonal regrowth. But if you have a better idea of the molecules and signals that facilitate the repair process naturally, then you might be able to enhance the function of the conduit, and that is exactly what we have been studying.’
‘We’ve known for a long time that dedifferentiation occurs but our findings show the full extent of this process,’ Parrinello explained. ‘It’s a reprogramming to a state that has properties of embryonic stem cells and not just a reversal to more immature Schwann cell type.
The study also reveals that the Schwann cells of the nerve bridge are different from other regions of the injured nerve in that they proliferate and invade more.
Parrinello described this finding as ‘very unexpected’ because it suggests that the cells adjust their characteristics depending on the environment in order to repair more efficiently. The studies also revealed that a specific molecule in the bridge, called TGFb, is important in driving this bridge-specific change.
Parrinello believes that her team’s findings could have wider physiological applications beyond simply improving nerve repair. Intriguingly, she says, the transcriptional profile of the bridge cells in the LMS study is reminiscent of cancer cells.
‘Understanding the signals that drive wound repair is relevant to cancer research because most solid tumours, are very similar to wounds,’ she adds. ‘So a better knowledge of how the signals work normally when they are well regulated could help to identify specific candidate molecules to block cancer.’
This study was a close collaboration between Dr Parrinello’s team and colleague Samuel Marguerat at the MRC Laboratory of Medical Sciences, who carried out analysis of the transcriptomic data. Prof. Alison Lloyd’s team at the MRC Laboratory for Molecular Cell biology, UCL also contributed to the work.
