Research published today provides novel understanding of what happens during gene regulation in eukaryotes, organisms that contain cells with genetic material contained within the nucleus. The surprising discovery from Dr Peter Sarkies, Head of Transgenerational Epigenetic Inheritance & Evolution, and his team, within the Epigenetics section at LMS, provides new knowledge about the pathways in human gene regulation and offers potential for targeting faulty gene regulation in cancer.
Peter’s group studies the evolution of epigenetic gene regulation, the ability of a gene to be switched on or off without the DNA/RNA being edited itself. Within organisms there are many processes for gene regulation, one of which is DNA methylation. In DNA methylation, genes called DNA methlytransferases add a chemical flag to Cytosine or C (one of the four bases in DNA A, G, C and T). This chemical flag often acts as a long-lasting signal to turn a gene off.
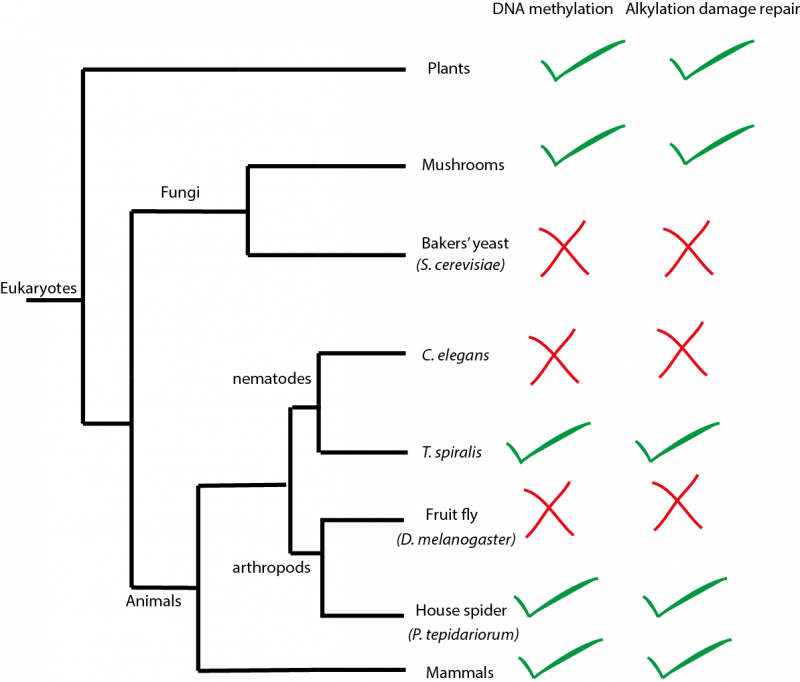
A simplified evolutionary tree showing some of the species of eukaryotes that the laboratory analysed. Whenever the DNA methylation system is present the DNA alkylation repair system is also present, indicating that the DNA methylation system might be the cause of the damage that the alkylation repair system usually repairs.
DNA methylation is an ancient mechanism to regulate genes found in diverse species of eukaryotes such as mammals, plants and fungi. It is essential for gene regulation in mammals, which will fail to develop normally without it. However, many species of eukaryotes have completely dispensed with DNA methylation during their evolution. These species include many well-known organisms used in research such as fruit flies, the nematode worm C. elegans and yeasts (see figure above). Interestingly, cancer cells in humans also often evolve to have strongly reduced levels of DNA methylation. However, we don’t understand why these changes in DNA methylation occur.
To start finding answers Peter and his team first looked at which other genes are present when the DNA methyltransferase genes that introduce DNA methylation are found, and are missing when DNA methyltransferases are lost. Surprisingly one of the clearest examples they found was a gene called ALKB2. ALKB2 is responsible for repairing DNA damaged by alkylation, a process which can lead to a mutation in DNA or a toxic block to the replication of DNA leading to cell death. Previously DNA alkylation was only known to occur when cells are exposed to toxic chemicals. The research team guessed that perhaps DNA methyltransferases genes might also introduce DNA alkylation damage into DNA. Working with Dr Petra Hajkova, Head of Reprogramming and Chromatin, they tested this idea and found that DNA methyltransferases do indeed damage DNA at the same time as carrying out their normal function in gene regulation.
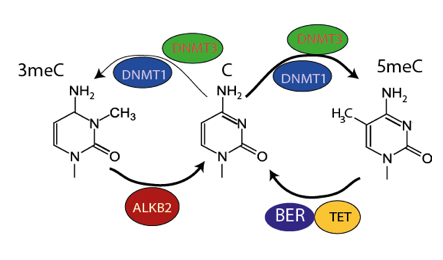
A model illustrating the danger of DNA methyltransferases. The predominant activity of DNA methyltransferases is to introduce gene regulatory 5 methyl cytosine into DNA, which can be converted back to cytosine by a DNA demethylase enzyme called TET. The LMS scientists also discovered that DNA methyltransferases occasionally introduces alkylation to cytosine in DNA, to create the toxic lesion 3 methyl cytosine. This requires the presence of a specialised repair enzyme called ALKB2 whenever DNA methyltransferases are present.
This unexpected discovery, that ALKB2 coevolves with DNA methyltransferase DNA methylation, provides conclusions backed by evidence that DNA methylation causes DNA damage, and that this damage is fixed by ALKB2. The toxic activity of DNA methyltransferases may explain why so many species have got rid of the process altogether and importantly, it might explain why cancer cells, which are very sensitive to DNA damage, have low levels of DNA methylation. The findings raise the possibility that increasing DNA methyltransferase activity might be a way to specifically target cancer cells for destruction, thus helping to treat the disease.
The full paper can be read here.
