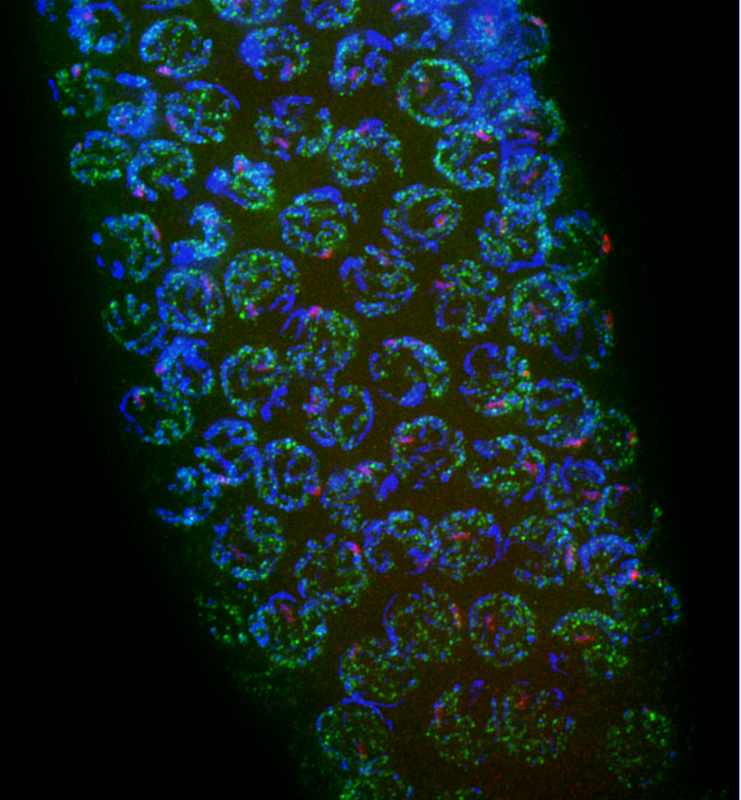
Binding of the transcription factor complex that controls piRNA transcription (PRDE-1, red) to the piRNA clusters in C.elegans germ cells. DNA has been stained with DAPI in blue, and the RNA polymerase II enzyme responsible for generating long mRNA transcripts can be seen in green.
“Treason is a matter of habit”. John le Carre was referring to spies when he wrote this but surprisingly the statement also applies to a large proportion of our own genomes. Our “enemies within” are known as Transposable Elements (TEs) or ‘jumping genes’: sequences of DNA that can selfishly copy themselves within the genome, potentially inserting themselves into essential genes and stopping those genes from functioning. One way in which cells fight back against these treacherous molecules is by generating piwi RNAs (piRNAs). piRNAs can bind to the RNA produced by TEs to stop them spreading. But where these piRNAs came from and how they were produced was still a bit of a mystery. Especially as the piRNA pathway is so diverse, even amongst closely related organisms, such as nematode worms.
Research published on 31 January in Developmental Cell by the Epigenetic Inheritance & Evolution group at the LMS, provides a new insight into how piRNAs are generated in nematodes. In particular, the role that chromatin and the transcriptional machinery play in piRNA biogenesis were highlighted. Chromatin is the complex of DNA and proteins that allow DNA to be packaged into a cell’s nucleus. The transcriptional machinery is the proteins that read the DNA sequence and convert it into an RNA message, or messenger RNA (mRNA) transcript.
The key finding of this research was that piRNA genes were clustered within repressed chromatin. These regions of the genome house genes that are usually silenced, but the piRNAs located here were still active. It also gave an insight into how the normal transcriptional machinery, that traditionally generates long mRNA transcripts, may have been subtly modified due to evolutionary pressure to create these short RNAs of 21-30 nucleotides in length that protect the genome.
These findings indicate how important the location of the piRNA genes in the genome is for their biogenesis. Given that the piRNA pathway is universal across species, this study also highlights how nematodes are different, even from some of their closest relatives.
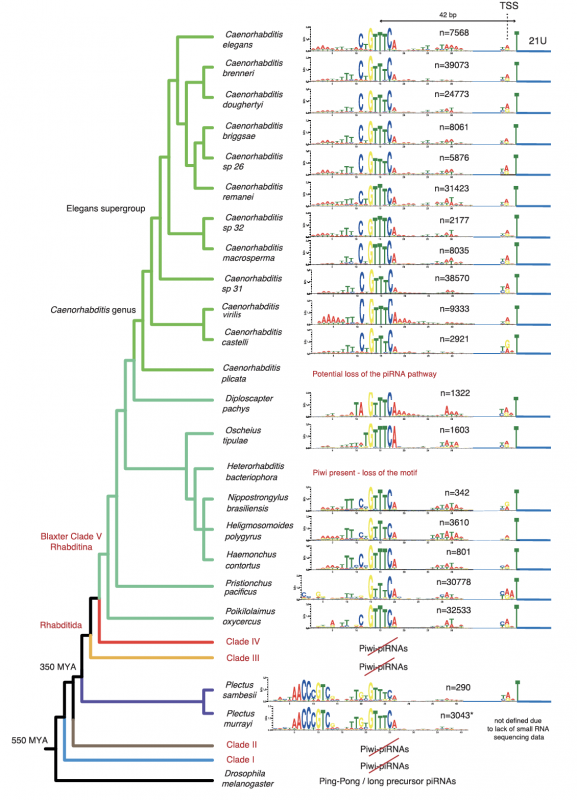
Evolutionary tree indicating all the similarities and differences in piRNA promoter structures between all the nematode species studied in this work. Read more about this image in Figure 1 of the publication.
Genome defence mechanisms need to evolve very rapidly to protect the DNA from TEs. To keep up, sometimes, the best path is to use what you already have in front of you and change it for your needs. The observation that the mechanisms for piRNA biogenesis are so varied amongst many nematodes, but also other model organisms like Drosophila melanogaster, highlights the intense pressure for these defence systems to stay ahead of the TEs and evolve rapidly.
Toni Beltran, PhD student and first author of this publication, had a unique approach to solve the question of the piRNA biogenesis question.

Toni Beltran, PhD student and first author
“From my background in biochemistry and an interest in RNA, I soon realised that to truly understand how cells work and to appreciate their complexity, I needed to apply computational approaches. When we realised that the piRNA genes were clustered within a 3.5Mb region of the genome we wanted to dive deeper into what features lay behind this organisation.”
The comparative genomics approach used by the team to answer the origin question had not been used before, and Toni Beltran said this about that aspect of their work:
“Many others in the field are focussed on generating new data, when in fact there is a lot to learn from just integrating what is already out there”.
Peter Sarkies, Head of the Epigenetic Inheritance and Evolution group at LMS and senior author
Peter Sarkies, senior author of the paper, shared the next steps for the research:
“Currently we just have the correlation between the chromatin state and the piRNA gene loci, so the next steps would be to better understand the mechanism behind how the chromatin stops the RNA polymerase from working.”
This publication also provided an insight into the evolution of piRNA biogenesis, which you can read more about in the blog post from senior author Peter Sarkies.
‘Comparative epigenomics reveals that RNA polymerase II pausing and chromatin domain organisation control nematode piRNA biogenesis’ was published in Developmental Cell on 31 January.

